참고 : Linear Models for the Prediction of Animal Breeding Values, 3rd Edtion. 9.3 Random Regression Model
임의 회귀 모형은 해를 구하고 나서도 해야할 일이 많다. 개체의 육종가 그래프를 그려 개체별로 비유기 동안 육종가가 어떻게 변하는지 살펴볼 수도 있다. 사실 모든 개체에 대해서 이렇게 그래프를 그려도 비교하기가 어렵기 때문에 사실상 할 일은 아니지만 그래도 개체의 일일 육종가를 구하여 그래프를 그리는 연습을 해 보자. 왜? 책에 그래프가 있으므로. 그리고 6일부터 310일까지의 일일 육종가를 구하면 305일 육종가가 된다. 아마도 이걸 비교하여 개체를 선발할 것이다. 어떤 프로그램을 사용하든 임의 회귀 모형의 해를 구했다고 가정하고 시작하자. 먼저 비유기 동안 분산의 변화부터 살펴보고나서 개체 육종가 그래프와 305일 육종가를 계산하자.
주의해야 할 것이 하나 있다. 예를 들어 유전평가를 할 때 DIM(days in milk, 분만후 비유일수)을 4에서 310을 했을 경우 즉 DIM 4에서 310까지의 르장드르 다항식을 사용하였을 경우 뒤의 모든 분석에서 그 다항식을 사용해야 한다. 예를 들어 305일 유지량을 계산하려면 DIM 6에서 DIM 310까지의 매일 매일 육종가를 더하는데 이때 DIM 6에서 310까지의 르장드르 다항식을 새로 만들어 계산하면 안된다. 유전 평가에서 사용했던 DIM 4에서 310의 르장드르 다항식 중 6에서 310까지만 추출해서 써야 한다. 책에는 설명이 안 되어 있고 해 보니 깨닫게 되는 중요한 사항이다.
여기서 르장드르 다항식(Legendre Polynomials)을 구하는 것이 중요한데 그것은 인터넷에서 복사한 것임을 밝힌다. 다음 사이트를 참조한다.
morotalab.org/Mrode2005/index.html
Mrode2005
morotalab.org
R 스크립트
# plotting the genetic and permanent environmental variance for DIM
# plotting the animals breeding values for DIM
## Given time points covariate and order of fit for Legendre polynomials, return matrix 'M' containing the polynomials of standardized time. 'M' is order t (number of time points) by k (order of Legendre polynomials).
## Arguments
## t: a vector of time, age or days
## n: order of polynomials
## tmax: max time (optional)
## tmin: min time (optional)
## Literature: Mrode, R.A. 2005. Linear Models for the Prediction of Animal Breeding Values. CAB International, Oxon, UK.
## Author: Gota Morota <morota at wisc dot edu>
## Created: 31-Mar-2010
## Last-Modified: 2-Apr-2010
## License: GPLv3 or later
`stdtime` <-
function(t, n, tmax, tmin){
if(missing(tmax)) {
tmax <- t[which.max(t)]
}
if(missing(tmin)) {
tmin <- t[which.min(t)]
}
N <- n+1
M <- matrix(0, nrow=length(t), ncol=N)
a <- -1 + 2*(t-tmin)/(tmax - tmin)
M[,1] <- 1
for (i in 2:N){
M[,i] <- a^(i-1)
}
return(M)
}
## Return coefficient matrix (lambda) of n-th order Legendre polynomials
## Arguments
## n: order of polynomials
## gengler: logical value. If TRUE, Genlger's scaling (1999) will be applied. If not specified, TRUE is assumed.
## Literatures
## Mrode, R.A. 2005. Linear Models for the Prediction of Animal Breeding Values. CAB International, Oxon, UK.
## Gengler, N. et. al. 1999. Estimation of (Co)variance Function Coefficients for Test Day Yield with a Expectation-Maximization Restricted Maximum Likelihood Algorithm. Journal of Dairy Science. 82
## Author: Gota Morota <morota at wisc dot edu>
## Create: 31-Mar-2010
## Last-Modified: 2-Apr-2010
## License: GPLv3 or later
`legendre` <-
function(n, gengler){
if (nargs()==1){
gengler <- TRUE
}
if (gengler != TRUE & gengler != FALSE){
gengler=TRUE
}
N <- n+1
L <- matrix(0,nrow=N, ncol=N)
for(i in (1:N)){
if(i==1){
L[i,i] <- 1
}
else if(i==2){
L[i,i] <- 1
}
else {
tmp <- L[i-1,]
tmp2 <- as.numeric()
tmp2 <- c(0,tmp[1:(N-1)])
L[i,] <- (1/(i-2+1))*( (2*(i-2) + 1)*tmp2 -(i-2)*L[i-2,] )
}
}
# Normalize
for (j in (1:N)){
L[j,] <- (sqrt( (2*(j-1)+1)/2) )*L[j,]
}
# Gengler (1999)
if (gengler==TRUE){
L <- sqrt(2)*L
}
return(L)
}
## example
## DIM <- c(4,38,72,106,140,174,208,242,276,310)
## order <- 4
## M <- stdtime(DIM, order)
## Lambda <- legendre(order, gengler=FALSE)
## Phi <- M%*%t(Lambda)
## Phi
# days in milk
dim = c(4:310)
head(dim)
tail(dim)
# order of animal regression coefficients
order = 2
# Mu : the matrix containing the polynomials of the standardized DIM values
M <- stdtime(dim, order)
head(M)
tail(M)
## Lambda : a matrix of order k containing the coefficients of Legendre polynomials.
Lambda <- legendre(order, gengler=FALSE)
head(Lambda)
tail(Lambda)
## Phi : Legendre polynomials of days in milk
Phi <- M %*% t(Lambda)
head(Phi)
tail(Phi)
# genetic variance and covariance
G = matrix(c( 3.297, 0.594, -1.381,
0.594, 0.921, -0.289,
-1.321, -0.289, 1.005 ), nrow = 3, byrow = TRUE)
G
P = matrix(c( 6.872, -0.254, -1.101,
-0.254, 3.171, 0.167,
-1.101, 0.167, 2.457 ), nrow = 3, byrow = TRUE)
P
# genetic variance for DIM i (vii)
fdim = 3
vii = matrix(rep(0, 3 * 307) , ncol = 3)
head(vii)
for (i in 1 : 307){
vii[i, 1] = fdim + i
vii[i, 2] = Phi[i,] %*% G %*% matrix(Phi[i,])
vii[i, 3] = Phi[i,] %*% P %*% matrix(Phi[i,])
}
head(vii)
tail(vii)
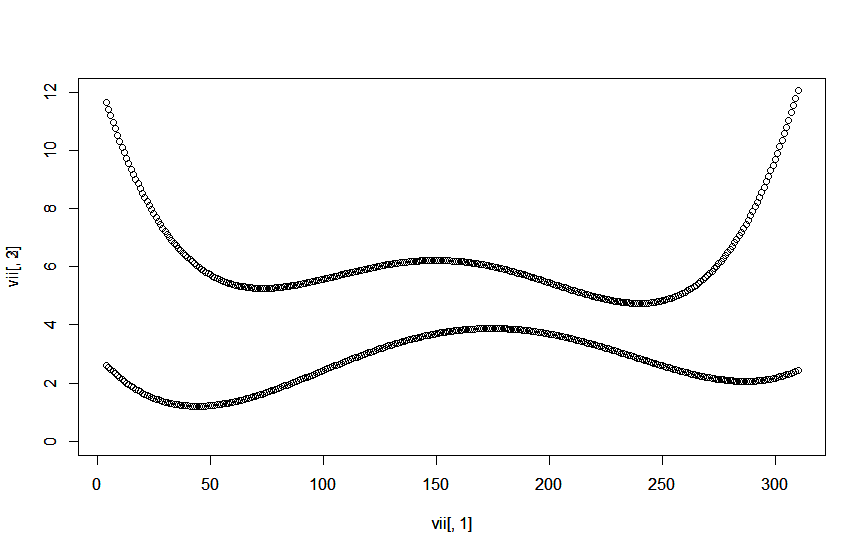
plot(vii[,1], vii[,2], ylim = c(0,12))
par(new = TRUE)
plot(vii[,1], vii[,3], ylim = c(0,12))
# breeding value of each animal for DIM i
# regression coefficients for animal effects
bvreg = matrix(c(
1, -0.0583, 0.0552, -0.0442,
2, -0.0728, -0.0305, -0.0244,
3, 0.1312, -0.0247, 0.0685,
4, 0.3445, 0.0063, -0.3164,
5, -0.4538, -0.052 , 0.2798,
6, -0.5486, 0.073 , 0.1946,
7, 0.8518, -0.0095, -0.3131,
8, 0.2208, 0.0127, -0.0174
), nrow = 8, byrow = TRUE)
bvreg
bvdim = matrix(rep(0, 9 * 307), ncol = 9)
head(bvdim)
for(i in 1 : 307){
bvdim[i, 1] = fdim + i
for(j in 1 : 8){
bvdim[i, j + 1] = Phi[i,] %*% matrix(bvreg[j, 2:4])
}
}
head(bvdim)
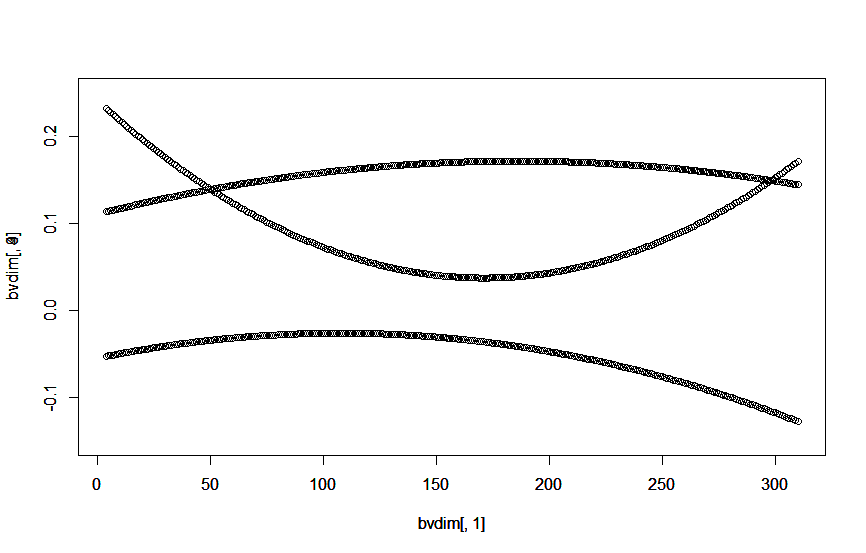
plot(bvdim[,1], bvdim[,3], ylim = c(-0.15,0.25))
par(new = TRUE)
plot(bvdim[,1], bvdim[,4], ylim = c(-0.15,0.25))
par(new = TRUE)
plot(bvdim[,1], bvdim[,9], ylim = c(-0.15,0.25))
# 305d breeding value
Phi2 = Phi[3 : 307,]
dim(Phi2)
t = colSums(Phi2)
t
bv_animal = rep(0, 8)
bv_animal
for(i in 1:8){
bv_animal[i] = t %*% matrix(bvreg[i, 2:4])
}
bv_animal
실행 결과
> # plotting the genetic and permanent environmental variance for DIM
> # plotting the animals breeding values for DIM
>
>
> ## Given time points covariate and order of fit for Legendre polynomials, return matrix 'M' containing the polynomials of standardized time. 'M' is order t (number of time points) by k (order of Legendre polynomials).
>
> ## Arguments
> ## t: a vector of time, age or days
> ## n: order of polynomials
> ## tmax: max time (optional)
> ## tmin: min time (optional)
>
> ## Literature: Mrode, R.A. 2005. Linear Models for the Prediction of Animal Breeding Values. CAB International, Oxon, UK.
>
> ## Author: Gota Morota <morota at wisc dot edu>
> ## Created: 31-Mar-2010
> ## Last-Modified: 2-Apr-2010
> ## License: GPLv3 or later
>
> `stdtime` <-
+ function(t, n, tmax, tmin){
+ if(missing(tmax)) {
+ tmax <- t[which.max(t)]
+ }
+ if(missing(tmin)) {
+ tmin <- t[which.min(t)]
+ }
+
+ N <- n+1
+ M <- matrix(0, nrow=length(t), ncol=N)
+ a <- -1 + 2*(t-tmin)/(tmax - tmin)
+ M[,1] <- 1
+
+ for (i in 2:N){
+ M[,i] <- a^(i-1)
+ }
+
+ return(M)
+ }
>
> ## Return coefficient matrix (lambda) of n-th order Legendre polynomials
>
> ## Arguments
> ## n: order of polynomials
> ## gengler: logical value. If TRUE, Genlger's scaling (1999) will be applied. If not specified, TRUE is assumed.
>
> ## Literatures
> ## Mrode, R.A. 2005. Linear Models for the Prediction of Animal Breeding Values. CAB International, Oxon, UK.
> ## Gengler, N. et. al. 1999. Estimation of (Co)variance Function Coefficients for Test Day Yield with a Expectation-Maximization Restricted Maximum Likelihood Algorithm. Journal of Dairy Science. 82
>
> ## Author: Gota Morota <morota at wisc dot edu>
> ## Create: 31-Mar-2010
> ## Last-Modified: 2-Apr-2010
> ## License: GPLv3 or later
>
> `legendre` <-
+ function(n, gengler){
+
+ if (nargs()==1){
+ gengler <- TRUE
+ }
+
+ if (gengler != TRUE & gengler != FALSE){
+ gengler=TRUE
+ }
+
+ N <- n+1
+ L <- matrix(0,nrow=N, ncol=N)
+
+ for(i in (1:N)){
+ if(i==1){
+ L[i,i] <- 1
+ }
+ else if(i==2){
+ L[i,i] <- 1
+ }
+ else {
+ tmp <- L[i-1,]
+ tmp2 <- as.numeric()
+ tmp2 <- c(0,tmp[1:(N-1)])
+ L[i,] <- (1/(i-2+1))*( (2*(i-2) + 1)*tmp2 -(i-2)*L[i-2,] )
+ }
+ }
+
+ # Normalize
+ for (j in (1:N)){
+ L[j,] <- (sqrt( (2*(j-1)+1)/2) )*L[j,]
+ }
+
+
+ # Gengler (1999)
+ if (gengler==TRUE){
+ L <- sqrt(2)*L
+ }
+
+ return(L)
+
+ }
>
> ## example
> ## DIM <- c(4,38,72,106,140,174,208,242,276,310)
> ## order <- 4
> ## M <- stdtime(DIM, order)
> ## Lambda <- legendre(order, gengler=FALSE)
> ## Phi <- M%*%t(Lambda)
> ## Phi
>
>
> # days in milk
> dim = c(4:310)
> head(dim)
[1] 4 5 6 7 8 9
> tail(dim)
[1] 305 306 307 308 309 310
>
> # order of animal regression coefficients
> order = 2
>
> # Mu : the matrix containing the polynomials of the standardized DIM values
> M <- stdtime(dim, order)
> head(M)
[,1] [,2] [,3]
[1,] 1 -1.0000000 1.0000000
[2,] 1 -0.9934641 0.9869708
[3,] 1 -0.9869281 0.9740271
[4,] 1 -0.9803922 0.9611688
[5,] 1 -0.9738562 0.9483959
[6,] 1 -0.9673203 0.9357085
> tail(M)
[,1] [,2] [,3]
[302,] 1 0.9673203 0.9357085
[303,] 1 0.9738562 0.9483959
[304,] 1 0.9803922 0.9611688
[305,] 1 0.9869281 0.9740271
[306,] 1 0.9934641 0.9869708
[307,] 1 1.0000000 1.0000000
>
> ## Lambda : a matrix of order k containing the coefficients of Legendre polynomials.
> Lambda <- legendre(order, gengler=FALSE)
> head(Lambda)
[,1] [,2] [,3]
[1,] 0.7071068 0.000000 0.000000
[2,] 0.0000000 1.224745 0.000000
[3,] -0.7905694 0.000000 2.371708
> tail(Lambda)
[,1] [,2] [,3]
[1,] 0.7071068 0.000000 0.000000
[2,] 0.0000000 1.224745 0.000000
[3,] -0.7905694 0.000000 2.371708
>
> ## Phi : Legendre polynomials of days in milk
> Phi <- M %*% t(Lambda)
> head(Phi)
[,1] [,2] [,3]
[1,] 0.7071068 -1.224745 1.581139
[2,] 0.7071068 -1.216740 1.550237
[3,] 0.7071068 -1.208735 1.519539
[4,] 0.7071068 -1.200730 1.489043
[5,] 0.7071068 -1.192725 1.458749
[6,] 0.7071068 -1.184721 1.428658
> tail(Phi)
[,1] [,2] [,3]
[302,] 0.7071068 1.184721 1.428658
[303,] 0.7071068 1.192725 1.458749
[304,] 0.7071068 1.200730 1.489043
[305,] 0.7071068 1.208735 1.519539
[306,] 0.7071068 1.216740 1.550237
[307,] 0.7071068 1.224745 1.581139
>
> # genetic variance and covariance
> G = matrix(c( 3.297, 0.594, -1.381,
+ 0.594, 0.921, -0.289,
+ -1.321, -0.289, 1.005 ), nrow = 3, byrow = TRUE)
> G
[,1] [,2] [,3]
[1,] 3.297 0.594 -1.381
[2,] 0.594 0.921 -0.289
[3,] -1.321 -0.289 1.005
>
> P = matrix(c( 6.872, -0.254, -1.101,
+ -0.254, 3.171, 0.167,
+ -1.101, 0.167, 2.457 ), nrow = 3, byrow = TRUE)
> P
[,1] [,2] [,3]
[1,] 6.872 -0.254 -1.101
[2,] -0.254 3.171 0.167
[3,] -1.101 0.167 2.457
>
> # genetic variance for DIM i (vii)
> fdim = 3
> vii = matrix(rep(0, 3 * 307) , ncol = 3)
> head(vii)
[,1] [,2] [,3]
[1,] 0 0 0
[2,] 0 0 0
[3,] 0 0 0
[4,] 0 0 0
[5,] 0 0 0
[6,] 0 0 0
>
> for (i in 1 : 307){
+ vii[i, 1] = fdim + i
+ vii[i, 2] = Phi[i,] %*% G %*% matrix(Phi[i,])
+ vii[i, 3] = Phi[i,] %*% P %*% matrix(Phi[i,])
+ }
> head(vii)
[,1] [,2] [,3]
[1,] 4 2.612026 11.66624
[2,] 5 2.533496 11.42854
[3,] 6 2.457661 11.19690
[4,] 7 2.384484 10.97121
[5,] 8 2.313922 10.75139
[6,] 9 2.245937 10.53735
> tail(vii)
[,1] [,2] [,3]
[302,] 305 2.279769 10.81685
[303,] 306 2.306494 11.05675
[304,] 307 2.334957 11.30292
[305,] 308 2.365192 11.55545
[306,] 309 2.397234 11.81442
[307,] 310 2.431118 12.07994
>
> plot(vii[,1], vii[,2], ylim = c(0,12))
> par(new = TRUE)
> plot(vii[,1], vii[,3], ylim = c(0,12))
>
>
> # breeding value of each animal for DIM i
> # regression coefficients for animal effects
> bvreg = matrix(c(
+ 1, -0.0583, 0.0552, -0.0442,
+ 2, -0.0728, -0.0305, -0.0244,
+ 3, 0.1312, -0.0247, 0.0685,
+ 4, 0.3445, 0.0063, -0.3164,
+ 5, -0.4538, -0.052 , 0.2798,
+ 6, -0.5486, 0.073 , 0.1946,
+ 7, 0.8518, -0.0095, -0.3131,
+ 8, 0.2208, 0.0127, -0.0174
+ ), nrow = 8, byrow = TRUE)
> bvreg
[,1] [,2] [,3] [,4]
[1,] 1 -0.0583 0.0552 -0.0442
[2,] 2 -0.0728 -0.0305 -0.0244
[3,] 3 0.1312 -0.0247 0.0685
[4,] 4 0.3445 0.0063 -0.3164
[5,] 5 -0.4538 -0.0520 0.2798
[6,] 6 -0.5486 0.0730 0.1946
[7,] 7 0.8518 -0.0095 -0.3131
[8,] 8 0.2208 0.0127 -0.0174
>
> bvdim = matrix(rep(0, 9 * 307), ncol = 9)
> head(bvdim)
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
[1,] 0 0 0 0 0 0 0 0 0
[2,] 0 0 0 0 0 0 0 0 0
[3,] 0 0 0 0 0 0 0 0 0
[4,] 0 0 0 0 0 0 0 0 0
[5,] 0 0 0 0 0 0 0 0 0
[6,] 0 0 0 0 0 0 0 0 0
>
> for(i in 1 : 307){
+ bvdim[i, 1] = fdim + i
+ for(j in 1 : 8){
+ bvdim[i, j + 1] = Phi[i,] %*% matrix(bvreg[j, 2:4])
+ }
+ }
> head(bvdim)
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
[1,] 4 -0.1787166 -0.05270244 0.2313316 -0.2643899 0.1852043 -0.1696355 0.1188941 0.1130631
[2,] 5 -0.1769089 -0.05219260 0.2290172 -0.2545623 0.1761419 -0.1750646 0.1284932 0.1137024
[3,] 6 -0.1751101 -0.05168770 0.2267166 -0.2447988 0.1671361 -0.1804542 0.1380290 0.1143383
[4,] 7 -0.1733203 -0.05118774 0.2244299 -0.2350994 0.1581870 -0.1858044 0.1475013 0.1149706
[5,] 8 -0.1715395 -0.05069272 0.2221570 -0.2254641 0.1492946 -0.1911152 0.1569101 0.1155993
[6,] 9 -0.1697676 -0.05020266 0.2198981 -0.2158929 0.1404590 -0.1963865 0.1662555 0.1162246
>
> plot(bvdim[,1], bvdim[,3], ylim = c(-0.15,0.25))
> par(new = TRUE)
> plot(bvdim[,1], bvdim[,4], ylim = c(-0.15,0.25))
> par(new = TRUE)
> plot(bvdim[,1], bvdim[,9], ylim = c(-0.15,0.25))
>
> # 305d breeding value
> Phi2 = Phi[3 : 307,]
> dim(Phi2)
[1] 305 3
> t = colSums(Phi2)
> t
[1] 215.667568 2.441485 -1.545070
>
> bv_animal = rep(0, 8)
> bv_animal
[1] 0 0 0 0 0 0 0 0
>
> for(i in 1:8){
+ bv_animal[i] = t %*% matrix(bvreg[i, 2:4])
+ }
>
> bv_animal
[1] -12.37036 -15.73736 28.12944 74.80172 -98.42921 -118.43767 184.16620 47.67729
>
일일 분산 그래프
일일 개체 육종가 그래프
X축 Y축 제목 넣고 예쁘게 하는 것은 필요할 때 하자.
'Animal Breeding > R for Genetic Evaluation' 카테고리의 다른 글
| Fixed Effect Model for SNP Effects, weighted analysis (0) | 2020.12.16 |
|---|---|
| Fixed Effect Model for SNP Effects, unweighted analysis (0) | 2020.12.16 |
| 다형질 모형의 차원 줄이기(Reduce the Dimension of Multivariate Models) (0) | 2020.09.29 |
| R을 이용한 일반적인 모형의 육종가 구하기 프로그래밍 (0) | 2020.08.25 |
| R을 이용한 다형질 개체 모형(unequal design matrices)의 육종가 구하기 프로그래밍 (0) | 2020.07.29 |