multiple trait animal model, same model and missing record
Linear Models for the Prediction of Animal Breeding Values, 3rd Edtion
Raphael Mrode
Example 5.2 p78
자료
4 1 4.5 0
5 2 2.9 5.0
6 2 3.9 6.8
7 1 3.5 6.0
8 1 5.0 7.5
9 2 4.0 0
mt02_data.txt 로 저장
혈통
1 0 0
2 0 0
3 0 0
4 1 0
5 3 2
6 1 2
7 4 5
8 3 6
9 7 0
mt02_pedi.txt 로 저장
자료와 혈통을 읽어 육종가를 구하는 R 프로그램이다.
신뢰도, 정확도, Standard error of prediction을 구한다.
육종가를 PA(parenta average), YD(yield devation), PC(progeny contribution)으로 구한다.
개체의 DYD(daughter yield deviation)을 구한다.
# multiple trait animal model, same model and missing record - Date : 2020-07-26
# Linear Models for the Prediction of Animal Breeding Values, 3rd Edtion
# Raphael Mrode
# Example 5.2 p78
# MASS packages
if (!("MASS" %in% installed.packages())) {
install.packages("MASS", dependencies = TRUE)
}
library(MASS)
# functions
# find the position in mixed model equation(lhs and rhs)
pos_mme <- function(trts, lvls, vals) {
pos = rep(0, length(vals))
for (i in 1:length(vals)) {
if (i == 1) {
pos[i] = trts * (vals[i] - 1) + 1
} else {
pos[i] = trts * sum(lvls[1:i - 1]) + trts * (vals[i] - 1) + 1
}
}
return(pos)
}
# make design matrix
desgn <- function(v) {
if (is.numeric(v)) {
va = v
mrow = length(va)
mcol = max(va)
}
if (is.character(v)) {
vf = factor(v)
# 각 수준의 인덱스 값을 저장
va = as.numeric(vf)
mrow = length(va)
mcol = length(levels(vf))
}
# 0으로 채워진 X 준비
X = matrix(data = c(0), nrow = mrow, ncol = mcol)
for (i in 1:mrow) {
ic = va[i]
X[i, ic] = 1
}
return(X)
}
# function to make inverse of numerator relationship matrix
ainv = function(pedi) {
n = nrow(pedi)
Ainv = matrix(c(0), nrow = n, ncol = n)
for (i in 1:n) {
animal = pedi[i, 1]
sire = pedi[i, 2]
dam = pedi[i, 3]
if (sire == 0 & dam == 0) {
# both parents unknown
alpha = 1
Ainv[animal, animal] = alpha + Ainv[animal, animal]
} else if (sire != 0 & dam == 0) {
# sire known
alpha = 4/3
Ainv[animal, animal] = alpha + Ainv[animal, animal]
Ainv[animal, sire] = -alpha/2 + Ainv[animal, sire]
Ainv[sire, animal] = -alpha/2 + Ainv[sire, animal]
Ainv[sire, sire] = alpha/4 + Ainv[sire, sire]
} else if (sire == 0 & dam != 0) {
# dam known
alpha = 4/3
Ainv[animal, animal] = alpha + Ainv[animal, animal]
Ainv[animal, dam] = -alpha/2 + Ainv[animal, dam]
Ainv[dam, animal] = -alpha/2 + Ainv[dam, animal]
Ainv[dam, dam] = alpha/4 + Ainv[dam, dam]
} else {
# both parents known
alpha = 2
Ainv[animal, animal] = alpha + Ainv[animal, animal]
Ainv[animal, sire] = -alpha/2 + Ainv[animal, sire]
Ainv[sire, animal] = -alpha/2 + Ainv[sire, animal]
Ainv[animal, dam] = -alpha/2 + Ainv[animal, dam]
Ainv[dam, animal] = -alpha/2 + Ainv[dam, animal]
Ainv[sire, sire] = alpha/4 + Ainv[sire, sire]
Ainv[sire, dam] = alpha/4 + Ainv[sire, dam]
Ainv[dam, sire] = alpha/4 + Ainv[dam, sire]
Ainv[dam, dam] = alpha/4 + Ainv[dam, dam]
}
}
return(Ainv)
}
# set working directory
#setwd(choose.dir())
setwd("D:/temp/05_multiple_traits_02_R")
# print working directory
getwd()
# no_of_trts
no_trts = 2
# list all possible combination of data
dtcombi0 = expand.grid(rep(list(0:1), no_trts))
dtcombi = dtcombi0[2:nrow(dtcombi0), ]
rownames(dtcombi) = NULL
# print
dtcombi
# prdigree and data
# read pedigree : animal sire dam
pedi = read.table("mt02_pedi.txt", header = FALSE, sep = "", col.names = c("animal", "sire", "dam"),
stringsAsFactors = FALSE)
pedi = pedi[order(pedi$animal), ]
# print
pedi
# read data : animal, dam, sex, weaning_weight
data = read.table("mt02_data.txt", header = FALSE, sep = "", col.names = c("animal", "sex", "wwg", "pwg"),
stringsAsFactors = FALSE)
# print
data
# number of data
no_data = nrow(data)
# print
no_data
# how many traits does animal have?
data2 = data.frame(data, dtsts = c(0))
data2$dtsts = ifelse(data2$wwg != 0, data2$dtsts + 1, data2$dtsts)
data2$dtsts = ifelse(data2$pwg != 0, data2$dtsts + 2, data2$dtsts)
# print
data2
# levels of animal and sex
lvls_ani = max(data2$animal)
lvls_sex = max(data2$sex)
# print
lvls_ani
lvls_sex
# variance component additive genetic
G = matrix(c(20, 18, 18, 40), 2, 2)
# residual
R = matrix(c(40, 11, 11, 30), 2, 2)
# print
G
R
# inverse of G
Gi = ginv(G)
# print
Gi
# inverse of R
Ri = array(rep(0, no_trts * no_trts * (2^no_trts - 1)), dim = c(no_trts, no_trts, (2^no_trts - 1)))
for (i in 1:(2^no_trts - 1)) {
R0 = R
R0[which(dtcombi[i, ] == 0), ] = 0
R0[, which(dtcombi[i, ] == 0)] = 0
Ri[, , i] = ginv(R0)
}
# print
Ri
# empty lhs
lhs = matrix(rep(0, (no_trts * (lvls_ani + lvls_sex))^2), nrow = no_trts * (lvls_ani + lvls_sex))
# print
dim(lhs)
lhs
# empty rhs
rhs = matrix(rep(0, (no_trts * (lvls_ani + lvls_sex))), nrow = no_trts * (lvls_ani + lvls_sex))
# print
dim(rhs)
rhs
# fill the MME
for (i in 1:no_data) {
#i = 1
pos = pos_mme(no_trts, c(lvls_sex, lvls_ani), c(data2$sex[i], data2$animal[i]))
for (j in 1:length(pos)) {
for (k in 1:length(pos)) {
rfirst = pos[j]
rlast = (pos[j] + no_trts - 1)
cfirst = pos[k]
clast = (pos[k] + no_trts - 1)
lhs[rfirst : rlast, cfirst : clast] = lhs[rfirst : rlast, cfirst : clast] + Ri[, , data2$dtsts[i]]
}
rhs[rfirst : rlast] = rhs[rfirst : rlast] + Ri[, , data2$dtsts[i]] %*% as.numeric(data2[i, 3:4])
}
}
# print lhs and rhs
lhs
rhs
# inverse matrix of NRM
ai = ainv(pedi)
# print
ai
# add ai to lhs
afirst = no_trts * lvls_sex + 1
alast = no_trts * (lvls_sex + lvls_ani)
lhs[afirst : alast, afirst : alast] = lhs[afirst : alast, afirst : alast] + ai %x% Gi
# print
lhs
# generalised inverse of lhs
gi_lhs = ginv(lhs)
# print
gi_lhs
# solution
sol = gi_lhs %*% rhs
# print
sol
# levels of fixed effect in traits 1
lvls_t1_f1 = rep(0, lvls_sex)
for ( i in 1 : lvls_sex){
if (i == 1){
lvls_t1_f1[i] = 1
} else {
lvls_t1_f1[i] = 1 + (i - 1) * no_trts
}
}
# print
lvls_t1_f1
# levels of fixed effect in traits 1
lvls_t2_f1 = lvls_t1_f1 + 1
# print
lvls_t2_f1
# levels of animal effect in traits 1
lvls_t1_ani = rep(0, lvls_ani)
for ( i in 1 : lvls_ani){
if (i == 1){
lvls_t1_ani[i] = 1
} else {
lvls_t1_ani[i] = 1 + (i - 1) * no_trts
}
}
lvls_t1_ani = lvls_t1_ani + no_trts * lvls_sex
# print
lvls_t1_ani
# levels of fixed effect in traits 1
lvls_t2_ani = lvls_t1_ani + 1
# print
lvls_t2_ani
# solutions for fixed effects and animal effects
sol_t1_f1 = as.matrix(sol[lvls_t1_f1])
sol_t2_f1 = as.matrix(sol[lvls_t2_f1])
sol_f1 = cbind(sol_t1_f1, sol_t2_f1)
# print
sol_f1
# breedinv value
sol_t1_bv = sol[lvls_t1_ani]
sol_t2_bv = sol[lvls_t2_ani]
sol_bv = cbind(sol_t1_bv, sol_t2_bv)
# print
sol_bv
# reliability(r2), accuracy(r), standard error of prediction(SEP)
# diagonal elements of the generalized inverse of LHS for animal equation
d_ani_t1 = diag(gi_lhs[lvls_t1_ani, lvls_t1_ani])
d_ani_t2 = diag(gi_lhs[lvls_t2_ani, lvls_t2_ani])
d_ani = cbind(d_ani_t1, d_ani_t2)
# print
d_ani
# reliability
rel = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
for (i in 1 : lvls_ani) {
rel[i, ] = 1 - d_ani[i, ]/diag(G)
}
# print
rel
# accuracy
acc = sqrt(rel)
# print
acc
# standard error of prediction(SEP)
sep = sqrt(d_ani)
# 확인
sep
# partitioning of breeding values
# yield deviation
yd1 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
# numerator of n2
a2 = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
# looping data
for (i in 1:no_data) {
yd1[data[i, 1], ] = as.matrix(data2[i, c(3, 4)] - sol_f1[data2[i, 2], ])
a2[, , data[i, 1]] = Ri[, , data2$dtsts[i]]
}
# print
yd1
a2
# Parents average, progeny contribution
# parents avearge
pa1 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
# progeny contribution numerator
pc0 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
# numerator of n3, denominator of progeny contribution
a3 = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
# numerator of n1
a1 = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
# looping pedi
for (i in 1 : lvls_ani) {
sire = pedi[i, 2]
dam = pedi[i, 3]
if (sire == 0 & dam == 0) {
# both parents unknown PA
a1[, , i] = 1 * Gi
} else if (sire != 0 & dam == 0) {
# 개체의 부만 알고 있을 경우
# PA
a1[, , i] = 4/3 * Gi
pa1[i, ] = sol_bv[sire, ]/2
# PC for sire
a3[, , sire] = a3[, , sire] + 0.5 * Gi * (2/3)
pc0[sire, ] = pc0[sire, ] + (0.5 * Gi * (2/3)) %*% (2 * sol_bv[i, ])
} else if (sire == 0 & dam != 0) {
# dam known
# PA
a1[, , i] = 4/3 * Gi
pa1[i, ] = sol_bv[dam, ]/2
# PC for dam
a3[, , dam] = a3[, , dam] + 0.5 * Gi * (2/3)
pc0[dam, ] = pc0[dam, ] + (0.5 * Gi * (2/3)) %*% (2 * sol_bv[i])
} else {
# both parents known
# PA
a1[, , i] = 2 * Gi
pa1[i, ] = (sol_bv[sire, ] + sol_bv[dam, ])/2
# PC for sire
a3[, , sire] = a3[, , sire] + 0.5 * Gi
pc0[sire, ] = pc0[sire, ] + (0.5 * Gi) %*% (2 * sol_bv[i, ] - sol_bv[dam, ])
# PC for dam
a3[, , dam] = a3[, , dam] + 0.5 * Gi
pc0[dam, ] = pc0[dam, ] + (0.5 * Gi) %*% (2 * sol_bv[i, ] - sol_bv[sire, ])
}
}
# print
a1
pa1
a3
pc0
# denominator of n1, n2, n3, diagonal of animals in LHS
n_de = a1 + a2 + a3
# print
n_de
# parents average fraction of breeding values
pa = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
for (i in 1 : lvls_ani) {
pa[i, ] = ginv(n_de[, , i]) %*% a1[, , i] %*% pa1[i, ]
}
# print
pa
# yield deviation fraction of breeding values
yd = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
for (i in 1 : lvls_ani) {
yd[i, ] = ginv(n_de[, , i]) %*% a2[, , i] %*% yd1[i, ]
}
# print
yd
# progeny contribution
pc1 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
for (i in 1 : lvls_ani) {
pc1[i, ] = ginv(a3[, , i]) %*% pc0[i, ]
}
pc1[is.nan(pc1) == TRUE] = 0
pc1
# progeny contribution fraction of breeding values
pc = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
for (i in 1 : lvls_ani) {
pc[i, ] = ginv(n_de[, , i]) %*% a3[, , i] %*% pc1[i, ]
}
# print
pc
# Progeny(Daughter) Yield Deviation(PYD, DYD)
# n2 of progeny
n2prog = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
for (i in 1 : lvls_ani) {
n2prog[, , i] = ginv((a1 + a2)[, , i]) %*% a2[, , i]
}
# print
n2prog
# numerator of dyd : summation of u of progeny * n2 of progeny * (2 * YD - bv of mate)
dyd_n = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
# denominator of dyd : summation of u of progeny * n2 of progeny
dyd_d = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
# looping pedi
for (i in 1 : lvls_ani) {
# i = 5
sire = pedi[i, 2]
dam = pedi[i, 3]
if (sire == 0 & dam == 0) {
# both parents unknown
} else if (sire != 0 & dam == 0) {
# 개체의 부만 알고 있을 경우
# dyd_n
dyd_n[sire, ] = dyd_n[sire, ] + (n2prog[, , i] * 2/3) %*% (2 * yd1[i, ])
# dyd_d
dyd_d[, , sire] = dyd_d[, , sire] + n2prog[, , i] * 2/3
} else if (sire == 0 & dam != 0) {
# dam known
# dyd_n
dyd_n[dam, ] = dyd_n[dam, ] + (n2prog[, , i] * 2/3) %*% (2 * yd1[i, ])
# dyd_d
dyd_d[, , dam] = dyd_d[, , dam] + n2prog[, , i] * 2/3
} else {
# both parents known
# dyd_n
dyd_n[sire, ] = dyd_n[sire, ] + n2prog[, , i] %*% (2 * yd1[i, ] - sol_bv[dam, ])
dyd_n[dam, ] = dyd_n[dam, ] + n2prog[, , i] %*% (2 * yd1[i, ] - sol_bv[sire, ])
# dyd_d
dyd_d[, , sire] = dyd_d[, , sire] + n2prog[, , i]
dyd_d[, , dam] = dyd_d[, , dam] + n2prog[, , i]
}
}
# print
dyd_n
dyd_d
# dyd
dyd = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
# looping pedi
for (i in 1 : lvls_ani) {
dyd[i, ] = ginv(dyd_d[, , i]) %*% dyd_n[i, ]
}
dyd[is.nan(dyd) == TRUE] = 0
# print
dyd
# breeding values and fractions
mt2_result = data.frame(animal = pedi[, 1], animal_bv = sol_bv, rel = rel, acc = acc, sep = sep, pa = pa,
yd = yd, pc = pc, sum_of_fr = pa + yd + pc, dyd = dyd)
# print
mt2_result
# 파일로 출력, 분리자는 ',', 따옴표 없고
output_filename = gsub("[ -]", "", paste("mt2_result_", Sys.Date(), ".csv"))
write.table(mt2_result, output_filename, sep = ",", row.names = FALSE, quote = FALSE)
다음은 실행 결과이다.
> # multiple trait animal model, same model and missing record - Date : 2020-07-26
>
> # Linear Models for the Prediction of Animal Breeding Values, 3rd Edtion
>
> # Raphael Mrode
>
> # Example 5.2 p78
>
> # MASS packages
> if (!("MASS" %in% installed.packages())) {
+ install.packages("MASS", dependencies = TRUE)
+ }
> library(MASS)
>
> # functions
>
> # find the position in mixed model equation(lhs and rhs)
> pos_mme <- function(trts, lvls, vals) {
+ pos = rep(0, length(vals))
+
+ for (i in 1:length(vals)) {
+ if (i == 1) {
+ pos[i] = trts * (vals[i] - 1) + 1
+ } else {
+ pos[i] = trts * lvls[i - 1] + trts * (vals[i] - 1) + 1
+ }
+ }
+
+ return(pos)
+ }
>
> # make design matrix
> desgn <- function(v) {
+ if (is.numeric(v)) {
+ va = v
+ mrow = length(va)
+ mcol = max(va)
+ }
+ if (is.character(v)) {
+ vf = factor(v)
+ # 각 수준의 인덱스 값을 저장
+ va = as.numeric(vf)
+ mrow = length(va)
+ mcol = length(levels(vf))
+ }
+
+ # 0으로 채워진 X 준비
+ X = matrix(data = c(0), nrow = mrow, ncol = mcol)
+
+ for (i in 1:mrow) {
+ ic = va[i]
+ X[i, ic] = 1
+ }
+ return(X)
+ }
>
> # function to make inverse of numerator relationship matrix
> ainv = function(pedi) {
+ n = nrow(pedi)
+ Ainv = matrix(c(0), nrow = n, ncol = n)
+
+ for (i in 1:n) {
+ animal = pedi[i, 1]
+ sire = pedi[i, 2]
+ dam = pedi[i, 3]
+
+ if (sire == 0 & dam == 0) {
+ # both parents unknown
+ alpha = 1
+ Ainv[animal, animal] = alpha + Ainv[animal, animal]
+ } else if (sire != 0 & dam == 0) {
+ # sire known
+ alpha = 4/3
+ Ainv[animal, animal] = alpha + Ainv[animal, animal]
+ Ainv[animal, sire] = -alpha/2 + Ainv[animal, sire]
+ Ainv[sire, animal] = -alpha/2 + Ainv[sire, animal]
+ Ainv[sire, sire] = alpha/4 + Ainv[sire, sire]
+ } else if (sire == 0 & dam != 0) {
+ # dam known
+ alpha = 4/3
+ Ainv[animal, animal] = alpha + Ainv[animal, animal]
+ Ainv[animal, dam] = -alpha/2 + Ainv[animal, dam]
+ Ainv[dam, animal] = -alpha/2 + Ainv[dam, animal]
+ Ainv[dam, dam] = alpha/4 + Ainv[dam, dam]
+ } else {
+ # both parents known
+ alpha = 2
+ Ainv[animal, animal] = alpha + Ainv[animal, animal]
+ Ainv[animal, sire] = -alpha/2 + Ainv[animal, sire]
+ Ainv[sire, animal] = -alpha/2 + Ainv[sire, animal]
+ Ainv[animal, dam] = -alpha/2 + Ainv[animal, dam]
+ Ainv[dam, animal] = -alpha/2 + Ainv[dam, animal]
+ Ainv[sire, sire] = alpha/4 + Ainv[sire, sire]
+ Ainv[sire, dam] = alpha/4 + Ainv[sire, dam]
+ Ainv[dam, sire] = alpha/4 + Ainv[dam, sire]
+ Ainv[dam, dam] = alpha/4 + Ainv[dam, dam]
+ }
+ }
+ return(Ainv)
+ }
>
> # set working directory
> #setwd(choose.dir())
> setwd("D:/temp/05_multiple_traits_02_R")
>
> # print working directory
> getwd()
[1] "D:/temp/05_multiple_traits_02_R"
>
> # no_of_trts
> no_trts = 2
>
> # list all possible combination of data
> dtcombi0 = expand.grid(rep(list(0:1), no_trts))
> dtcombi = dtcombi0[2:nrow(dtcombi0), ]
> rownames(dtcombi) = NULL
>
> # print
> dtcombi
Var1 Var2
1 1 0
2 0 1
3 1 1
>
> # prdigree and data
>
> # read pedigree : animal sire dam
> pedi = read.table("mt02_pedi.txt", header = FALSE, sep = "", col.names = c("animal", "sire", "dam"),
+ stringsAsFactors = FALSE)
> pedi = pedi[order(pedi$animal), ]
>
> # print
> pedi
animal sire dam
1 1 0 0
2 2 0 0
3 3 0 0
4 4 1 0
5 5 3 2
6 6 1 2
7 7 4 5
8 8 3 6
9 9 7 0
>
> # read data : animal, dam, sex, weaning_weight
> data = read.table("mt02_data.txt", header = FALSE, sep = "", col.names = c("animal", "sex", "wwg", "pwg"),
+ stringsAsFactors = FALSE)
>
> # print
> data
animal sex wwg pwg
1 4 1 4.5 0.0
2 5 2 2.9 5.0
3 6 2 3.9 6.8
4 7 1 3.5 6.0
5 8 1 5.0 7.5
6 9 2 4.0 0.0
>
> # number of data
> no_data = nrow(data)
>
> # print
> no_data
[1] 6
>
> # how many traits does animal have?
> data2 = data.frame(data, dtsts = c(0))
> data2$dtsts = ifelse(data2$wwg != 0, data2$dtsts + 1, data2$dtsts)
> data2$dtsts = ifelse(data2$pwg != 0, data2$dtsts + 2, data2$dtsts)
>
> # print
> data2
animal sex wwg pwg dtsts
1 4 1 4.5 0.0 1
2 5 2 2.9 5.0 3
3 6 2 3.9 6.8 3
4 7 1 3.5 6.0 3
5 8 1 5.0 7.5 3
6 9 2 4.0 0.0 1
>
> # levels of animal and sex
> lvls_ani = max(data2$animal)
> lvls_sex = max(data2$sex)
>
> # print
> lvls_ani
[1] 9
> lvls_sex
[1] 2
>
> # variance component additive genetic
> G = matrix(c(20, 18, 18, 40), 2, 2)
> # residual
> R = matrix(c(40, 11, 11, 30), 2, 2)
>
> # print
> G
[,1] [,2]
[1,] 20 18
[2,] 18 40
> R
[,1] [,2]
[1,] 40 11
[2,] 11 30
>
> # inverse of G
> Gi = ginv(G)
>
> # print
> Gi
[,1] [,2]
[1,] 0.084 -0.038
[2,] -0.038 0.042
>
> # inverse of R
> Ri = array(rep(0, no_trts * no_trts * (2^no_trts - 1)), dim = c(no_trts, no_trts, (2^no_trts - 1)))
>
> for (i in 1:(2^no_trts - 1)) {
+ R0 = R
+ R0[which(dtcombi[i, ] == 0), ] = 0
+ R0[, which(dtcombi[i, ] == 0)] = 0
+ Ri[, , i] = ginv(R0)
+ }
>
> # print
> Ri
, , 1
[,1] [,2]
[1,] 0.025 0
[2,] 0.000 0
, , 2
[,1] [,2]
[1,] 0 0.000
[2,] 0 0.033
, , 3
[,1] [,2]
[1,] 0.028 -0.010
[2,] -0.010 0.037
>
> # empty lhs
> lhs = matrix(rep(0, (no_trts * (lvls_ani + lvls_sex))^2), nrow = no_trts * (lvls_ani + lvls_sex))
>
> # print
> dim(lhs)
[1] 22 22
> lhs
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16] [,17] [,18] [,19] [,20] [,21] [,22]
[1,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[2,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[3,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[4,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[5,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[6,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[7,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[8,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[9,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[10,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[11,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[12,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[13,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[14,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[15,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[16,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[17,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[18,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[19,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[20,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[21,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[22,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
>
> # empty rhs
> rhs = matrix(rep(0, (no_trts * (lvls_ani + lvls_sex))), nrow = no_trts * (lvls_ani + lvls_sex))
>
> # print
> dim(rhs)
[1] 22 1
> rhs
[,1]
[1,] 0
[2,] 0
[3,] 0
[4,] 0
[5,] 0
[6,] 0
[7,] 0
[8,] 0
[9,] 0
[10,] 0
[11,] 0
[12,] 0
[13,] 0
[14,] 0
[15,] 0
[16,] 0
[17,] 0
[18,] 0
[19,] 0
[20,] 0
[21,] 0
[22,] 0
>
> # fill the MME
> for (i in 1:no_data) {
+ #i = 1
+ pos = pos_mme(no_trts, c(lvls_sex, lvls_ani), c(data2$sex[i], data2$animal[i]))
+
+ for (j in 1:length(pos)) {
+ for (k in 1:length(pos)) {
+ rfirst = pos[j]
+ rlast = (pos[j] + no_trts - 1)
+ cfirst = pos[k]
+ clast = (pos[k] + no_trts - 1)
+
+ lhs[rfirst : rlast, cfirst : clast] = lhs[rfirst : rlast, cfirst : clast] + Ri[, , data2$dtsts[i]]
+ }
+ rhs[rfirst : rlast] = rhs[rfirst : rlast] + Ri[, , data2$dtsts[i]] %*% as.numeric(data2[i, 3:4])
+ }
+ }
>
> # print lhs and rhs
> lhs
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16] [,17] [,18] [,19] [,20] [,21] [,22]
[1,] 0.081 -0.020 0.000 0.000 0 0 0 0 0 0 0.025 0 0.000 0.000 0.000 0.000 0.028 -0.010 0.028 -0.010 0.000 0
[2,] -0.020 0.074 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 -0.010 0.037 -0.010 0.037 0.000 0
[3,] 0.000 0.000 0.081 -0.020 0 0 0 0 0 0 0.000 0 0.028 -0.010 0.028 -0.010 0.000 0.000 0.000 0.000 0.025 0
[4,] 0.000 0.000 -0.020 0.074 0 0 0 0 0 0 0.000 0 -0.010 0.037 -0.010 0.037 0.000 0.000 0.000 0.000 0.000 0
[5,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[6,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[7,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[8,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[9,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[10,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[11,] 0.025 0.000 0.000 0.000 0 0 0 0 0 0 0.025 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[12,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[13,] 0.000 0.000 0.028 -0.010 0 0 0 0 0 0 0.000 0 0.028 -0.010 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[14,] 0.000 0.000 -0.010 0.037 0 0 0 0 0 0 0.000 0 -0.010 0.037 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
[15,] 0.000 0.000 0.028 -0.010 0 0 0 0 0 0 0.000 0 0.000 0.000 0.028 -0.010 0.000 0.000 0.000 0.000 0.000 0
[16,] 0.000 0.000 -0.010 0.037 0 0 0 0 0 0 0.000 0 0.000 0.000 -0.010 0.037 0.000 0.000 0.000 0.000 0.000 0
[17,] 0.028 -0.010 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.028 -0.010 0.000 0.000 0.000 0
[18,] -0.010 0.037 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 -0.010 0.037 0.000 0.000 0.000 0
[19,] 0.028 -0.010 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.028 -0.010 0.000 0
[20,] -0.010 0.037 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 -0.010 0.037 0.000 0
[21,] 0.000 0.000 0.025 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.025 0
[22,] 0.000 0.000 0.000 0.000 0 0 0 0 0 0 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0
> rhs
[,1]
[1,] 0.211
[2,] 0.414
[3,] 0.169
[4,] 0.368
[5,] 0.000
[6,] 0.000
[7,] 0.000
[8,] 0.000
[9,] 0.000
[10,] 0.000
[11,] 0.112
[12,] 0.000
[13,] 0.030
[14,] 0.156
[15,] 0.039
[16,] 0.212
[17,] 0.036
[18,] 0.187
[19,] 0.063
[20,] 0.227
[21,] 0.100
[22,] 0.000
>
> # inverse matrix of NRM
> ai = ainv(pedi)
>
> # print
> ai
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
[1,] 1.83 0.5 0.0 -0.67 0.0 -1.0 0.00 0 0.00
[2,] 0.50 2.0 0.5 0.00 -1.0 -1.0 0.00 0 0.00
[3,] 0.00 0.5 2.0 0.00 -1.0 0.5 0.00 -1 0.00
[4,] -0.67 0.0 0.0 1.83 0.5 0.0 -1.00 0 0.00
[5,] 0.00 -1.0 -1.0 0.50 2.5 0.0 -1.00 0 0.00
[6,] -1.00 -1.0 0.5 0.00 0.0 2.5 0.00 -1 0.00
[7,] 0.00 0.0 0.0 -1.00 -1.0 0.0 2.33 0 -0.67
[8,] 0.00 0.0 -1.0 0.00 0.0 -1.0 0.00 2 0.00
[9,] 0.00 0.0 0.0 0.00 0.0 0.0 -0.67 0 1.33
>
> # add ai to lhs
> afirst = no_trts * lvls_sex + 1
> alast = no_trts * (lvls_sex + lvls_ani)
> lhs[afirst : alast, afirst : alast] = lhs[afirst : alast, afirst : alast] + ai %x% Gi
>
> # print
> lhs
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16] [,17] [,18] [,19] [,20] [,21] [,22]
[1,] 0.081 -0.020 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.025 0.000 0.000 0.000 0.000 0.000 0.028 -0.010 0.028 -0.010 0.000 0.000
[2,] -0.020 0.074 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 -0.010 0.037 -0.010 0.037 0.000 0.000
[3,] 0.000 0.000 0.081 -0.020 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.028 -0.010 0.028 -0.010 0.000 0.000 0.000 0.000 0.025 0.000
[4,] 0.000 0.000 -0.020 0.074 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 -0.010 0.037 -0.010 0.037 0.000 0.000 0.000 0.000 0.000 0.000
[5,] 0.000 0.000 0.000 0.000 0.154 -0.069 0.042 -0.019 0.000 0.000 -0.056 0.025 0.000 0.000 -0.084 0.038 0.000 0.000 0.000 0.000 0.000 0.000
[6,] 0.000 0.000 0.000 0.000 -0.069 0.077 -0.019 0.021 0.000 0.000 0.025 -0.028 0.000 0.000 0.038 -0.042 0.000 0.000 0.000 0.000 0.000 0.000
[7,] 0.000 0.000 0.000 0.000 0.042 -0.019 0.168 -0.076 0.042 -0.019 0.000 0.000 -0.084 0.038 -0.084 0.038 0.000 0.000 0.000 0.000 0.000 0.000
[8,] 0.000 0.000 0.000 0.000 -0.019 0.021 -0.076 0.084 -0.019 0.021 0.000 0.000 0.038 -0.042 0.038 -0.042 0.000 0.000 0.000 0.000 0.000 0.000
[9,] 0.000 0.000 0.000 0.000 0.000 0.000 0.042 -0.019 0.168 -0.076 0.000 0.000 -0.084 0.038 0.042 -0.019 0.000 0.000 -0.084 0.038 0.000 0.000
[10,] 0.000 0.000 0.000 0.000 0.000 0.000 -0.019 0.021 -0.076 0.084 0.000 0.000 0.038 -0.042 -0.019 0.021 0.000 0.000 0.038 -0.042 0.000 0.000
[11,] 0.025 0.000 0.000 0.000 -0.056 0.025 0.000 0.000 0.000 0.000 0.179 -0.069 0.042 -0.019 0.000 0.000 -0.084 0.038 0.000 0.000 0.000 0.000
[12,] 0.000 0.000 0.000 0.000 0.025 -0.028 0.000 0.000 0.000 0.000 -0.069 0.077 -0.019 0.021 0.000 0.000 0.038 -0.042 0.000 0.000 0.000 0.000
[13,] 0.000 0.000 0.028 -0.010 0.000 0.000 -0.084 0.038 -0.084 0.038 0.042 -0.019 0.238 -0.105 0.000 0.000 -0.084 0.038 0.000 0.000 0.000 0.000
[14,] 0.000 0.000 -0.010 0.037 0.000 0.000 0.038 -0.042 0.038 -0.042 -0.019 0.021 -0.105 0.142 0.000 0.000 0.038 -0.042 0.000 0.000 0.000 0.000
[15,] 0.000 0.000 0.028 -0.010 -0.084 0.038 -0.084 0.038 0.042 -0.019 0.000 0.000 0.000 0.000 0.238 -0.105 0.000 0.000 -0.084 0.038 0.000 0.000
[16,] 0.000 0.000 -0.010 0.037 0.038 -0.042 0.038 -0.042 -0.019 0.021 0.000 0.000 0.000 0.000 -0.105 0.142 0.000 0.000 0.038 -0.042 0.000 0.000
[17,] 0.028 -0.010 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 -0.084 0.038 -0.084 0.038 0.000 0.000 0.224 -0.098 0.000 0.000 -0.056 0.025
[18,] -0.010 0.037 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.038 -0.042 0.038 -0.042 0.000 0.000 -0.098 0.135 0.000 0.000 0.025 -0.028
[19,] 0.028 -0.010 0.000 0.000 0.000 0.000 0.000 0.000 -0.084 0.038 0.000 0.000 0.000 0.000 -0.084 0.038 0.000 0.000 0.196 -0.086 0.000 0.000
[20,] -0.010 0.037 0.000 0.000 0.000 0.000 0.000 0.000 0.038 -0.042 0.000 0.000 0.000 0.000 0.038 -0.042 0.000 0.000 -0.086 0.121 0.000 0.000
[21,] 0.000 0.000 0.025 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 -0.056 0.025 0.000 0.000 0.137 -0.050
[22,] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.025 -0.028 0.000 0.000 -0.050 0.056
>
> # generalised inverse of lhs
> gi_lhs = ginv(lhs)
>
> # print
> gi_lhs
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16] [,17] [,18] [,19] [,20] [,21] [,22]
[1,] 23.8 13.1 6.1 5.6 -6.518 -5.796 -3.285 -2.956 -5.26 -4.88 -10.54 -9.34 -5.9 -5.4 -6.56 -5.9 -11.3 -10.0 -9.6 -8.8 -5.8 -5.1
[2,] 13.1 38.4 6.1 15.4 -5.248 -10.861 -3.700 -9.280 -5.64 -13.84 -7.53 -14.27 -6.4 -15.9 -6.46 -15.0 -10.7 -24.4 -10.0 -24.0 -5.6 -12.4
[3,] 6.1 6.1 22.5 12.0 -4.457 -4.020 -7.196 -6.477 -3.84 -3.50 -3.93 -3.45 -9.4 -8.4 -9.15 -8.3 -7.8 -6.9 -6.5 -6.0 -9.0 -8.0
[4,] 5.6 15.4 12.0 37.6 -4.397 -9.923 -7.567 -18.710 -3.69 -9.18 -3.39 -6.15 -9.6 -23.5 -9.58 -23.4 -7.0 -15.4 -6.4 -15.9 -5.8 -9.8
[5,] -6.5 -5.2 -4.5 -4.4 18.567 16.171 0.049 0.044 1.47 1.90 8.71 7.50 1.9 2.5 8.26 6.7 5.6 5.2 5.2 4.7 3.3 3.0
[6,] -5.8 -10.9 -4.0 -9.9 16.171 36.405 0.024 0.021 1.90 3.78 7.55 17.54 2.5 5.1 6.63 15.1 5.2 11.2 4.7 10.0 3.0 6.0
[7,] -3.3 -3.7 -7.2 -7.6 0.049 0.024 19.055 17.149 -0.56 -0.45 0.91 0.76 8.8 8.0 9.01 8.1 4.9 4.3 4.1 3.8 3.7 3.3
[8,] -3.0 -9.3 -6.5 -18.7 0.044 0.021 17.149 39.234 -0.50 -0.40 0.82 0.68 8.0 19.1 8.11 19.2 4.4 9.8 3.7 9.3 3.4 6.0
[9,] -5.3 -5.6 -3.8 -3.7 1.466 1.901 -0.561 -0.505 17.93 15.39 2.14 2.46 7.6 6.0 0.98 1.6 5.2 4.9 8.5 7.3 2.9 2.7
[10,] -4.9 -13.8 -3.5 -9.2 1.902 3.777 -0.450 -0.405 15.39 34.99 2.41 4.09 6.1 14.6 1.66 4.0 4.9 11.2 7.3 17.2 2.8 5.8
[11,] -10.5 -7.5 -3.9 -3.4 8.710 7.550 0.913 0.821 2.14 2.41 16.94 14.81 2.1 2.2 5.13 4.4 9.5 8.1 5.2 5.2 4.5 3.9
[12,] -9.3 -14.3 -3.5 -6.1 7.505 17.544 0.759 0.683 2.46 4.09 14.81 35.43 2.2 3.3 4.26 8.9 8.0 17.4 5.2 9.7 3.9 8.5
[13,] -5.9 -6.4 -9.4 -9.6 1.857 2.458 8.845 7.960 7.59 6.09 2.12 2.24 16.0 13.2 6.41 7.0 8.8 7.4 6.8 6.5 5.7 4.9
[14,] -5.4 -15.9 -8.4 -23.5 2.459 5.077 7.974 19.076 6.05 14.58 2.24 3.30 13.2 31.3 7.00 16.6 7.5 16.9 6.4 15.7 5.0 9.6
[15,] -6.6 -6.5 -9.2 -9.6 8.258 6.625 9.009 8.108 0.98 1.66 5.13 4.26 6.4 7.0 16.22 13.4 6.4 6.3 8.2 7.0 4.8 4.6
[16,] -5.9 -15.0 -8.3 -23.4 6.666 15.057 8.112 19.201 1.59 4.00 4.38 8.89 7.0 16.6 13.36 31.3 6.3 13.9 6.9 16.6 4.6 8.2
[17,] -11.3 -10.7 -7.8 -7.0 5.614 5.168 4.877 4.389 5.18 4.95 9.51 8.01 8.8 7.5 6.39 6.3 17.2 14.2 7.2 7.8 8.4 6.9
[18,] -10.0 -24.4 -6.9 -15.4 5.194 11.222 4.295 9.816 4.87 11.16 8.12 17.37 7.4 16.9 6.26 13.9 14.2 32.0 7.6 17.3 7.0 15.9
[19,] -9.6 -10.0 -6.5 -6.4 5.229 4.669 4.065 3.659 8.48 7.30 5.18 5.20 6.8 6.4 8.17 6.9 7.2 7.6 16.3 13.5 4.4 4.5
[20,] -8.8 -24.0 -6.0 -15.9 4.700 10.018 3.758 9.332 7.28 17.20 5.17 9.67 6.5 15.7 7.05 16.6 7.8 17.3 13.5 31.7 4.5 9.1
[21,] -5.8 -5.6 -9.0 -5.8 3.257 2.976 3.736 3.362 2.93 2.75 4.53 3.85 5.7 5.0 4.82 4.6 8.4 7.0 4.4 4.5 16.4 14.5
[22,] -5.1 -12.4 -8.0 -9.8 3.002 5.964 3.316 5.959 2.74 5.83 3.86 8.55 4.9 9.6 4.59 8.2 6.9 15.9 4.5 9.1 14.5 35.8
>
> # solution
> sol = gi_lhs %*% rhs
>
> # print
> sol
[,1]
[1,] 4.367
[2,] 6.834
[3,] 3.657
[4,] 6.007
[5,] 0.130
[6,] 0.266
[7,] -0.084
[8,] -0.075
[9,] -0.098
[10,] -0.194
[11,] 0.007
[12,] 0.016
[13,] -0.343
[14,] -0.555
[15,] 0.192
[16,] 0.440
[17,] -0.308
[18,] -0.483
[19,] 0.201
[20,] 0.349
[21,] -0.018
[22,] -0.119
>
> # levels of fixed effect in traits 1
> lvls_t1_f1 = rep(0, lvls_sex)
> for ( i in 1 : lvls_sex){
+ if (i == 1){
+ lvls_t1_f1[i] = 1
+ } else {
+ lvls_t1_f1[i] = 1 + (i - 1) * no_trts
+ }
+ }
>
> # print
> lvls_t1_f1
[1] 1 3
>
> # levels of fixed effect in traits 1
> lvls_t2_f1 = lvls_t1_f1 + 1
>
> # print
> lvls_t2_f1
[1] 2 4
>
> # levels of animal effect in traits 1
> lvls_t1_ani = rep(0, lvls_ani)
> for ( i in 1 : lvls_ani){
+ if (i == 1){
+ lvls_t1_ani[i] = 1
+ } else {
+ lvls_t1_ani[i] = 1 + (i - 1) * no_trts
+ }
+ }
> lvls_t1_ani = lvls_t1_ani + no_trts * lvls_sex
>
> # print
> lvls_t1_ani
[1] 5 7 9 11 13 15 17 19 21
>
> # levels of fixed effect in traits 1
> lvls_t2_ani = lvls_t1_ani + 1
>
> # print
> lvls_t2_ani
[1] 6 8 10 12 14 16 18 20 22
>
> # solutions for fixed effects and animal effects
> sol_t1_f1 = as.matrix(sol[lvls_t1_f1])
> sol_t2_f1 = as.matrix(sol[lvls_t2_f1])
> sol_f1 = cbind(sol_t1_f1, sol_t2_f1)
>
> # print
> sol_f1
[,1] [,2]
[1,] 4.4 6.8
[2,] 3.7 6.0
>
> # breedinv value
> sol_t1_bv = sol[lvls_t1_ani]
> sol_t2_bv = sol[lvls_t2_ani]
> sol_bv = cbind(sol_t1_bv, sol_t2_bv)
>
> # print
> sol_bv
sol_t1_bv sol_t2_bv
[1,] 0.130 0.266
[2,] -0.084 -0.075
[3,] -0.098 -0.194
[4,] 0.007 0.016
[5,] -0.343 -0.555
[6,] 0.192 0.440
[7,] -0.308 -0.483
[8,] 0.201 0.349
[9,] -0.018 -0.119
>
> # reliability(r2), accuracy(r), standard error of prediction(SEP)
>
> # diagonal elements of the generalized inverse of LHS for animal equation
> d_ani_t1 = diag(gi_lhs[lvls_t1_ani, lvls_t1_ani])
> d_ani_t2 = diag(gi_lhs[lvls_t2_ani, lvls_t2_ani])
> d_ani = cbind(d_ani_t1, d_ani_t2)
>
> # print
> d_ani
d_ani_t1 d_ani_t2
[1,] 19 36
[2,] 19 39
[3,] 18 35
[4,] 17 35
[5,] 16 31
[6,] 16 31
[7,] 17 32
[8,] 16 32
[9,] 16 36
>
> # reliability
> rel = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
>
> for (i in 1 : lvls_ani) {
+ rel[i, ] = 1 - d_ani[i, ]/diag(G)
+ }
>
> # print
> rel
[,1] [,2]
[1,] 0.072 0.090
[2,] 0.047 0.019
[3,] 0.103 0.125
[4,] 0.153 0.114
[5,] 0.200 0.217
[6,] 0.189 0.218
[7,] 0.141 0.200
[8,] 0.187 0.208
[9,] 0.180 0.106
>
> # accuracy
> acc = sqrt(rel)
>
> # print
> acc
[,1] [,2]
[1,] 0.27 0.30
[2,] 0.22 0.14
[3,] 0.32 0.35
[4,] 0.39 0.34
[5,] 0.45 0.47
[6,] 0.43 0.47
[7,] 0.38 0.45
[8,] 0.43 0.46
[9,] 0.42 0.33
>
> # standard error of prediction(SEP)
> sep = sqrt(d_ani)
>
> # 확인
> sep
d_ani_t1 d_ani_t2
[1,] 4.3 6.0
[2,] 4.4 6.3
[3,] 4.2 5.9
[4,] 4.1 6.0
[5,] 4.0 5.6
[6,] 4.0 5.6
[7,] 4.1 5.7
[8,] 4.0 5.6
[9,] 4.1 6.0
>
> # partitioning of breeding values
>
> # yield deviation
> yd1 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
>
> # numerator of n2
> a2 = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
>
> # looping data
> for (i in 1:no_data) {
+ yd1[data[i, 1], ] = as.matrix(data2[i, c(3, 4)] - sol_f1[data2[i, 2], ])
+
+ a2[, , data[i, 1]] = Ri[, , data2$dtsts[i]]
+ }
>
> # print
> yd1
[,1] [,2]
[1,] 0.00 0.00
[2,] 0.00 0.00
[3,] 0.00 0.00
[4,] 0.13 -6.83
[5,] -0.76 -1.01
[6,] 0.24 0.79
[7,] -0.87 -0.83
[8,] 0.63 0.67
[9,] 0.34 -6.01
> a2
, , 1
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 2
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 3
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 4
[,1] [,2]
[1,] 0.025 0
[2,] 0.000 0
, , 5
[,1] [,2]
[1,] 0.028 -0.010
[2,] -0.010 0.037
, , 6
[,1] [,2]
[1,] 0.028 -0.010
[2,] -0.010 0.037
, , 7
[,1] [,2]
[1,] 0.028 -0.010
[2,] -0.010 0.037
, , 8
[,1] [,2]
[1,] 0.028 -0.010
[2,] -0.010 0.037
, , 9
[,1] [,2]
[1,] 0.025 0
[2,] 0.000 0
>
> # Parents average, progeny contribution
>
> # parents avearge
> pa1 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
>
> # progeny contribution numerator
> pc0 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
>
> # numerator of n3, denominator of progeny contribution
> a3 = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
>
> # numerator of n1
> a1 = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
>
>
> # looping pedi
> for (i in 1 : lvls_ani) {
+
+ sire = pedi[i, 2]
+ dam = pedi[i, 3]
+
+ if (sire == 0 & dam == 0) {
+ # both parents unknown PA
+ a1[, , i] = 1 * Gi
+
+ } else if (sire != 0 & dam == 0) {
+ # 개체의 부만 알고 있을 경우
+
+ # PA
+ a1[, , i] = 4/3 * Gi
+ pa1[i, ] = sol_bv[sire, ]/2
+
+ # PC for sire
+ a3[, , sire] = a3[, , sire] + 0.5 * Gi * (2/3)
+ pc0[sire, ] = pc0[sire, ] + (0.5 * Gi * (2/3)) %*% (2 * sol_bv[i, ])
+
+ } else if (sire == 0 & dam != 0) {
+ # dam known
+
+ # PA
+ a1[, , i] = 4/3 * Gi
+ pa1[i, ] = sol_bv[dam, ]/2
+
+ # PC for dam
+ a3[, , dam] = a3[, , dam] + 0.5 * Gi * (2/3)
+ pc0[dam, ] = pc0[dam, ] + (0.5 * Gi * (2/3)) %*% (2 * sol_bv[i])
+
+ } else {
+ # both parents known
+
+ # PA
+ a1[, , i] = 2 * Gi
+ pa1[i, ] = (sol_bv[sire, ] + sol_bv[dam, ])/2
+
+ # PC for sire
+ a3[, , sire] = a3[, , sire] + 0.5 * Gi
+ pc0[sire, ] = pc0[sire, ] + (0.5 * Gi) %*% (2 * sol_bv[i, ] - sol_bv[dam, ])
+
+ # PC for dam
+ a3[, , dam] = a3[, , dam] + 0.5 * Gi
+ pc0[dam, ] = pc0[dam, ] + (0.5 * Gi) %*% (2 * sol_bv[i, ] - sol_bv[sire, ])
+
+ }
+ }
>
> # print
> a1
, , 1
[,1] [,2]
[1,] 0.084 -0.038
[2,] -0.038 0.042
, , 2
[,1] [,2]
[1,] 0.084 -0.038
[2,] -0.038 0.042
, , 3
[,1] [,2]
[1,] 0.084 -0.038
[2,] -0.038 0.042
, , 4
[,1] [,2]
[1,] 0.11 -0.050
[2,] -0.05 0.056
, , 5
[,1] [,2]
[1,] 0.168 -0.076
[2,] -0.076 0.084
, , 6
[,1] [,2]
[1,] 0.168 -0.076
[2,] -0.076 0.084
, , 7
[,1] [,2]
[1,] 0.168 -0.076
[2,] -0.076 0.084
, , 8
[,1] [,2]
[1,] 0.168 -0.076
[2,] -0.076 0.084
, , 9
[,1] [,2]
[1,] 0.11 -0.050
[2,] -0.05 0.056
> pa1
[,1] [,2]
[1,] 0.000 0.000
[2,] 0.000 0.000
[3,] 0.000 0.000
[4,] 0.065 0.133
[5,] -0.091 -0.135
[6,] 0.023 0.095
[7,] -0.168 -0.269
[8,] 0.047 0.123
[9,] -0.154 -0.241
> a3
, , 1
[,1] [,2]
[1,] 0.070 -0.032
[2,] -0.032 0.035
, , 2
[,1] [,2]
[1,] 0.084 -0.038
[2,] -0.038 0.042
, , 3
[,1] [,2]
[1,] 0.084 -0.038
[2,] -0.038 0.042
, , 4
[,1] [,2]
[1,] 0.042 -0.019
[2,] -0.019 0.021
, , 5
[,1] [,2]
[1,] 0.042 -0.019
[2,] -0.019 0.021
, , 6
[,1] [,2]
[1,] 0.042 -0.019
[2,] -0.019 0.021
, , 7
[,1] [,2]
[1,] 0.028 -0.013
[2,] -0.013 0.014
, , 8
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 9
[,1] [,2]
[1,] 0 0
[2,] 0 0
> pc0
[,1] [,2]
[1,] 0.0015 1.2e-02
[2,] -0.0084 -4.1e-16
[3,] -0.0018 -8.9e-03
[4,] -0.0037 -3.5e-03
[5,] -0.0076 -8.8e-03
[6,] 0.0041 9.3e-03
[7,] 0.0020 -2.9e-03
[8,] 0.0000 0.0e+00
[9,] 0.0000 0.0e+00
>
> # denominator of n1, n2, n3, diagonal of animals in LHS
> n_de = a1 + a2 + a3
>
> # print
> n_de
, , 1
[,1] [,2]
[1,] 0.154 -0.069
[2,] -0.069 0.077
, , 2
[,1] [,2]
[1,] 0.168 -0.076
[2,] -0.076 0.084
, , 3
[,1] [,2]
[1,] 0.168 -0.076
[2,] -0.076 0.084
, , 4
[,1] [,2]
[1,] 0.179 -0.069
[2,] -0.069 0.077
, , 5
[,1] [,2]
[1,] 0.24 -0.10
[2,] -0.10 0.14
, , 6
[,1] [,2]
[1,] 0.24 -0.10
[2,] -0.10 0.14
, , 7
[,1] [,2]
[1,] 0.224 -0.098
[2,] -0.098 0.135
, , 8
[,1] [,2]
[1,] 0.196 -0.086
[2,] -0.086 0.121
, , 9
[,1] [,2]
[1,] 0.14 -0.050
[2,] -0.05 0.056
>
> # parents average fraction of breeding values
> pa = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
> for (i in 1 : lvls_ani) {
+ pa[i, ] = ginv(n_de[, , i]) %*% a1[, , i] %*% pa1[i, ]
+ }
>
> # print
> pa
[,1] [,2]
[1,] 0.000 0.000
[2,] 0.000 0.000
[3,] 0.000 0.000
[4,] 0.037 0.088
[5,] -0.052 -0.070
[6,] 0.008 0.050
[7,] -0.099 -0.146
[8,] 0.025 0.074
[9,] -0.112 -0.204
>
> # yield deviation fraction of breeding values
> yd = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
> for (i in 1 : lvls_ani) {
+ yd[i, ] = ginv(n_de[, , i]) %*% a2[, , i] %*% yd1[i, ]
+ }
>
> # print
> yd
[,1] [,2]
[1,] 0.000 0.000
[2,] 0.000 0.000
[3,] 0.000 0.000
[4,] 0.029 0.026
[5,] -0.203 -0.358
[6,] 0.115 0.274
[7,] -0.208 -0.315
[8,] 0.176 0.275
[9,] 0.094 0.084
>
> # progeny contribution
> pc1 = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
> for (i in 1 : lvls_ani) {
+ pc1[i, ] = ginv(a3[, , i]) %*% pc0[i, ]
+ }
> pc1[is.nan(pc1) == TRUE] = 0
> pc1
[,1] [,2]
[1,] 0.286 0.59
[2,] -0.167 -0.15
[3,] -0.196 -0.39
[4,] -0.274 -0.41
[5,] -0.623 -0.98
[6,] 0.499 0.89
[7,] -0.037 -0.24
[8,] 0.000 0.00
[9,] 0.000 0.00
>
> # progeny contribution fraction of breeding values
> pc = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
> for (i in 1 : lvls_ani) {
+ pc[i, ] = ginv(n_de[, , i]) %*% a3[, , i] %*% pc1[i, ]
+ }
>
> # print
> pc
[,1] [,2]
[1,] 0.12982 0.266
[2,] -0.08361 -0.075
[3,] -0.09807 -0.194
[4,] -0.05861 -0.098
[5,] -0.08802 -0.127
[6,] 0.06827 0.116
[7,] -0.00079 -0.022
[8,] 0.00000 0.000
[9,] 0.00000 0.000
>
> # Progeny(Daughter) Yield Deviation(PYD, DYD)
>
> # n2 of progeny
> n2prog = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
> for (i in 1 : lvls_ani) {
+ n2prog[, , i] = ginv((a1 + a2)[, , i]) %*% a2[, , i]
+ }
>
> # print
> n2prog
, , 1
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 2
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 3
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 4
[,1] [,2]
[1,] 0.27 0
[2,] 0.25 0
, , 5
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 6
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 7
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 8
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 9
[,1] [,2]
[1,] 0.27 0
[2,] 0.25 0
>
> # numerator of dyd : summation of u of progeny * n2 of progeny * (2 * YD - bv of mate)
> dyd_n = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
> # denominator of dyd : summation of u of progeny * n2 of progeny
> dyd_d = array(rep(0, no_trts * no_trts * lvls_ani), dim = c(no_trts, no_trts, lvls_ani))
>
> # looping pedi
> for (i in 1 : lvls_ani) {
+ # i = 5
+ sire = pedi[i, 2]
+ dam = pedi[i, 3]
+
+ if (sire == 0 & dam == 0) {
+ # both parents unknown
+
+ } else if (sire != 0 & dam == 0) {
+ # 개체의 부만 알고 있을 경우
+
+ # dyd_n
+ dyd_n[sire, ] = dyd_n[sire, ] + (n2prog[, , i] * 2/3) %*% (2 * yd1[i, ])
+ # dyd_d
+ dyd_d[, , sire] = dyd_d[, , sire] + n2prog[, , i] * 2/3
+
+ } else if (sire == 0 & dam != 0) {
+ # dam known
+
+ # dyd_n
+ dyd_n[dam, ] = dyd_n[dam, ] + (n2prog[, , i] * 2/3) %*% (2 * yd1[i, ])
+ # dyd_d
+ dyd_d[, , dam] = dyd_d[, , dam] + n2prog[, , i] * 2/3
+
+ } else {
+ # both parents known
+
+ # dyd_n
+ dyd_n[sire, ] = dyd_n[sire, ] + n2prog[, , i] %*% (2 * yd1[i, ] - sol_bv[dam, ])
+ dyd_n[dam, ] = dyd_n[dam, ] + n2prog[, , i] %*% (2 * yd1[i, ] - sol_bv[sire, ])
+
+ # dyd_d
+ dyd_d[, , sire] = dyd_d[, , sire] + n2prog[, , i]
+ dyd_d[, , dam] = dyd_d[, , dam] + n2prog[, , i]
+
+ }
+ }
>
> # print
> dyd_n
[,1] [,2]
[1,] 0.33 0.71
[2,] -0.22 -0.22
[3,] -0.18 -0.42
[4,] -0.34 -0.47
[5,] -0.47 -0.70
[6,] 0.39 0.63
[7,] 0.12 0.11
[8,] 0.00 0.00
[9,] 0.00 0.00
> dyd_d
, , 1
[,1] [,2]
[1,] 0.33 0.12
[2,] 0.19 0.39
, , 2
[,1] [,2]
[1,] 0.305 0.24
[2,] 0.048 0.78
, , 3
[,1] [,2]
[1,] 0.305 0.24
[2,] 0.048 0.78
, , 4
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 5
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 6
[,1] [,2]
[1,] 0.152 0.12
[2,] 0.024 0.39
, , 7
[,1] [,2]
[1,] 0.18 0
[2,] 0.16 0
, , 8
[,1] [,2]
[1,] 0 0
[2,] 0 0
, , 9
[,1] [,2]
[1,] 0 0
[2,] 0 0
>
> # dyd
> dyd = matrix(rep(0, lvls_ani * no_trts), ncol = no_trts)
>
> # looping pedi
> for (i in 1 : lvls_ani) {
+ dyd[i, ] = ginv(dyd_d[, , i]) %*% dyd_n[i, ]
+ }
> dyd[is.nan(dyd) == TRUE] = 0
>
> # print
> dyd
[,1] [,2]
[1,] 0.43 1.60
[2,] -0.53 -0.25
[3,] -0.18 -0.52
[4,] -1.39 -1.11
[5,] -1.74 -1.68
[6,] 1.36 1.53
[7,] 0.69 0.00
[8,] 0.00 0.00
[9,] 0.00 0.00
>
> # breeding values and fractions
> mt2_result = data.frame(animal = pedi[, 1], animal_bv = sol_bv, rel = rel, acc = acc, sep = sep, pa = pa,
+ yd = yd, pc = pc, sum_of_fr = pa + yd + pc, dyd = dyd)
>
> # print
> mt2_result
animal animal_bv.sol_t1_bv animal_bv.sol_t2_bv rel.1 rel.2 acc.1 acc.2 sep.d_ani_t1 sep.d_ani_t2 pa.1 pa.2 yd.1 yd.2 pc.1 pc.2 sum_of_fr.1
1 1 0.130 0.266 0.072 0.090 0.27 0.30 4.3 6.0 0.000 0.000 0.000 0.000 0.12982 0.266 0.130
2 2 -0.084 -0.075 0.047 0.019 0.22 0.14 4.4 6.3 0.000 0.000 0.000 0.000 -0.08361 -0.075 -0.084
3 3 -0.098 -0.194 0.103 0.125 0.32 0.35 4.2 5.9 0.000 0.000 0.000 0.000 -0.09807 -0.194 -0.098
4 4 0.007 0.016 0.153 0.114 0.39 0.34 4.1 6.0 0.037 0.088 0.029 0.026 -0.05861 -0.098 0.007
5 5 -0.343 -0.555 0.200 0.217 0.45 0.47 4.0 5.6 -0.052 -0.070 -0.203 -0.358 -0.08802 -0.127 -0.343
6 6 0.192 0.440 0.189 0.218 0.43 0.47 4.0 5.6 0.008 0.050 0.115 0.274 0.06827 0.116 0.192
7 7 -0.308 -0.483 0.141 0.200 0.38 0.45 4.1 5.7 -0.099 -0.146 -0.208 -0.315 -0.00079 -0.022 -0.308
8 8 0.201 0.349 0.187 0.208 0.43 0.46 4.0 5.6 0.025 0.074 0.176 0.275 0.00000 0.000 0.201
9 9 -0.018 -0.119 0.180 0.106 0.42 0.33 4.1 6.0 -0.112 -0.204 0.094 0.084 0.00000 0.000 -0.018
sum_of_fr.2 dyd.1 dyd.2
1 0.266 0.43 1.60
2 -0.075 -0.53 -0.25
3 -0.194 -0.18 -0.52
4 0.016 -1.39 -1.11
5 -0.555 -1.74 -1.68
6 0.440 1.36 1.53
7 -0.483 0.69 0.00
8 0.349 0.00 0.00
9 -0.119 0.00 0.00
>
> # 파일로 출력, 분리자는 ',', 따옴표 없고
> output_filename = gsub("[ -]", "", paste("mt2_result_", Sys.Date(), ".csv"))
> write.table(mt2_result, output_filename, sep = ",", row.names = FALSE, quote = FALSE)
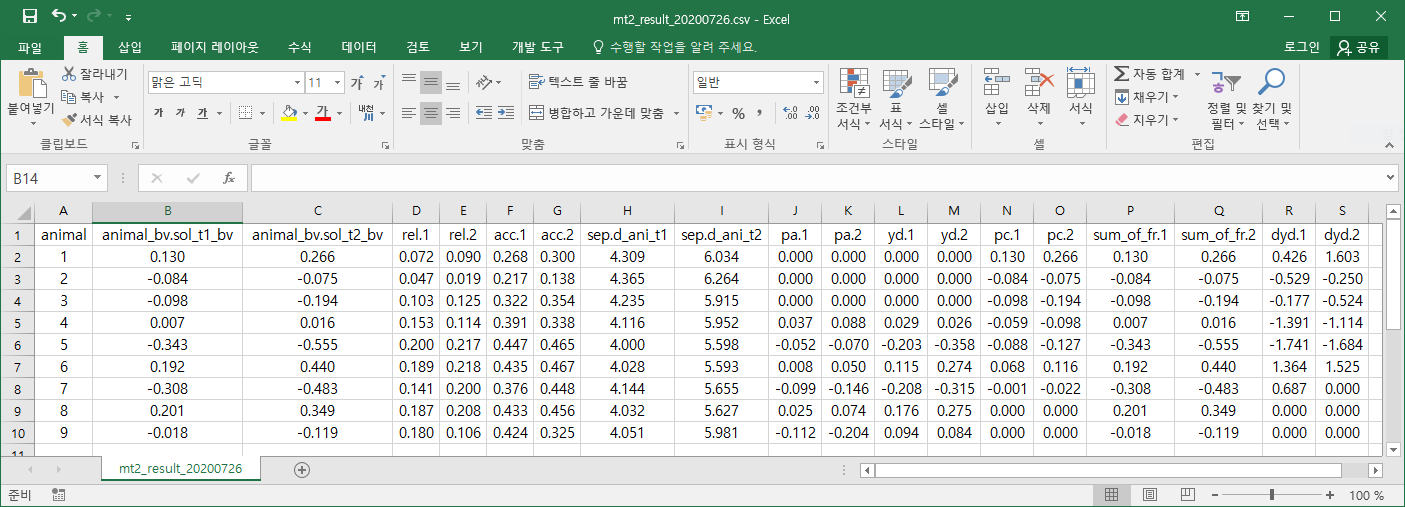
출력 파일(mt2_result_20200723.csv)은 다음과 같다.
animal,animal_bv.sol_t1_bv,animal_bv.sol_t2_bv,rel.1,rel.2,acc.1,acc.2,sep.d_ani_t1,sep.d_ani_t2,pa.1,pa.2,yd.1,yd.2,pc.1,pc.2,sum_of_fr.1,sum_of_fr.2,dyd.1,dyd.2
1,0.129823830715541,0.266245867684148,0.0716308723768238,0.0898698568225488,0.267639444732692,0.299783016234324,4.3089885765065,6.03367265660792,0,0,0,0,0.12982383071554,0.266245867684148,0.12982383071554,0.266245867684148,0.425630149209032,1.6034276646541
2,-0.0836108051833632,-0.0752497246650299,0.0472677904320411,0.0191434551249767,0.217411569223078,0.138359875415442,4.36516256184798,6.2637258716359,0,0,0,0,-0.0836108051833641,-0.0752497246650325,-0.0836108051833641,-0.0752497246650325,-0.529046133710049,-0.25000119324945
3,-0.0980748544120003,-0.194040392874268,0.103425883888995,0.125227278344733,0.321598948830674,0.353874664739838,4.23455810235497,5.91531139215939,0,0,0,0,-0.0980748544119998,-0.194040392874261,-0.0980748544119998,-0.194040392874261,-0.17737262403059,-0.523780147010833
4,0.0070204624237542,0.0155735319963907,0.152981516122118,0.114272108052331,0.391128516119853,0.33804157740185,4.11586803451685,5.95223619137436,0.0370925230615831,0.0877121507700289,0.0285364540447902,0.0256828086403112,-0.058608514682622,-0.0978214274139516,0.00702046242375134,0.0155735319963885,-1.39078799459603,-1.1142944367354
5,-0.342871767652598,-0.554506919015954,0.199940867464958,0.216657919742212,0.447147478428492,0.465465272326747,4.00014782860594,5.59764979346792,-0.0520194635641462,-0.0696089987726521,-0.202833831789998,-0.357877306170488,-0.0880184722984567,-0.127020614072812,-0.3428717676526,-0.554506919015952,-1.74068022467238,-1.68437488774775
6,0.191524645437639,0.440110207090828,0.188816172313118,0.217983462221048,0.434529829025716,0.466886990845802,4.02786252915087,5.59291172030795,0.00800973820069506,0.0500753419923305,0.115246263693708,0.274344181926019,0.0682686435432385,0.11569068317249,0.191524645437642,0.440110207090839,1.36441509216338,1.52523903712291
7,-0.308189084752417,-0.482966574707401,0.141437372423211,0.200445576913419,0.376081603409701,0.447711488476026,4.14382100862667,5.65527867778974,-0.099144837939897,-0.145827359673472,-0.208256444037528,-0.315253106749821,-0.000787802774987632,-0.0218861082841027,-0.308189084752413,-0.482966574707395,0.686828354441722,0
8,0.200678978955736,0.348856374480332,0.187187667563252,0.208313886948288,0.432651901143693,0.456414161643006,4.03190360112131,5.6273834525531,0.0249578986836898,0.0738802077728371,0.175721080272048,0.274976166707489,0,0,0.200678978955738,0.348856374480327,0,0
9,-0.018410346122461,-0.119367510725329,0.179508661013015,0.105647480996723,0.423684624470861,0.32503458430869,4.05090443971957,5.98114543880443,-0.112068758091788,-0.203660081497722,0.0936584119693256,0.0842925707723931,0,0,-0.0184103461224622,-0.119367510725329,0,0
csv 파일이므로 엑셀에서 열면 다음과 같다.
'Animal Breeding > R for Genetic Evaluation' 카테고리의 다른 글
| R을 이용한 일반적인 모형의 육종가 구하기 프로그래밍 (0) | 2020.08.25 |
|---|---|
| R을 이용한 다형질 개체 모형(unequal design matrices)의 육종가 구하기 프로그래밍 (0) | 2020.07.29 |
| R을 이용한 다형질 개체 모형(same model and no missing record)의 육종가 구하기 프로그래밍 (0) | 2020.07.03 |
| R을 이용하여 다형질 개체 모형(multiple trait animal model)에 대한 blupf90 분석 결과 정리하기 (0) | 2020.07.02 |
| R을 이용한 공통 환경 효과 모형(Common Environmental Effect Model)의 육종가 구하기 프로그래밍 (0) | 2020.07.01 |