# Linear Models for the Prediction of Animal Breeding Values, 3rd Edition.
# Raphael Mrode
# Example 11.5 p189
간단한 모델 설명은 다음 포스팅을 참고한다.
SNP-BLUP Model with Polygenic Effects(unweighted analysis)
# Linear Models for the Prediction of Animal Breeding Values, 3rd Edition. # Raphael Mrode # Example 11.5 p189 SNP effect로 모든 상가적 유전 분산을 설명할 수 없을 수도 있다. 그래서 설명..
bhpark.tistory.com
Data
13 0 0 1 558 9 0.00179211 1.3571429 -0.3571429 0.2857143 0.2857143 -0.2857143 -1.2142857 -0.1428571 0.07142857 -0.1428571 1.2142857
14 0 0 1 722 13.4 0.00138504 0.3571429 -0.3571429 -0.7142857 -0.7142857 -0.2857143 0.7857143 -0.1428571 0.07142857 -0.1428571 -0.7857143
15 13 4 1 300 12.7 0.00333333 0.3571429 0.6428571 1.2857143 0.2857143 0.7142857 -1.2142857 -0.1428571 0.07142857 -0.1428571 1.2142857
16 15 2 1 73 15.4 0.01369863 -0.6428571 -0.3571429 1.2857143 0.2857143 -0.2857143 -0.2142857 -0.1428571 0.07142857 0.8571429 0.2142857
17 15 5 1 52 5.9 0.01923077 -0.6428571 0.6428571 0.2857143 1.2857143 -0.2857143 -1.2142857 -0.1428571 0.07142857 -0.1428571 1.2142857
18 14 6 1 87 7.7 0.01149425 0.3571429 0.6428571 -0.7142857 0.2857143 -0.2857143 0.7857143 -0.1428571 0.07142857 0.8571429 0.2142857
19 14 9 1 64 10.2 0.01562500 -0.6428571 -0.3571429 0.2857143 0.2857143 -0.2857143 0.7857143 -0.1428571 0.07142857 0.8571429 -0.7857143
20 14 9 1 103 4.8 0.00970874 -0.6428571 0.6428571 0.2857143 -0.7142857 -0.2857143 -0.2142857 -0.1428571 0.07142857 0.8571429 -0.7857143
Pedigree
1 0 0
2 0 0
3 0 0
4 0 0
5 0 0
6 0 0
7 0 0
8 0 0
9 0 0
10 0 0
11 0 0
12 0 0
13 0 0
14 0 0
15 13 4
16 15 2
17 15 5
18 14 6
19 14 9
20 14 9
21 1 3
22 14 8
23 14 11
24 14 10
25 14 7
26 14 12
Renumf90을 위한 파라미터 파일
# Parameter file for program renf90; it is translated to parameter
# file for BLUPF90 family programs.
DATAFILE
snp_data2.txt
TRAITS
6
FIELDS_PASSED TO OUTPUT
WEIGHT(S)
RESIDUAL_VARIANCE
245
EFFECT
4 cross alpha
EFFECT
1 cross alpha
RANDOM
animal
FILE
snp_pedi2.txt
FILE_POS
1 2 3
PED_DEPTH
0
(CO)VARIANCES
3.5241
EFFECT
4 cross alpha
RANDOM
diagonal
RANDOM_REGRESSION
data
RR_POSITION
8 9 10 11 12 13 14 15 16 17
(CO)VARIANCES
8.963969 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 8.963969 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.00 8.963969 0.00 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 8.963969 0.00 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 8.963969 0.00 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 8.963969 0.00 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 8.963969 0.00 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 8.963969 0.00 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 8.963969 0.00
0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 8.963969
OPTION solv_method FSPAK
Renumf90 실행 화면
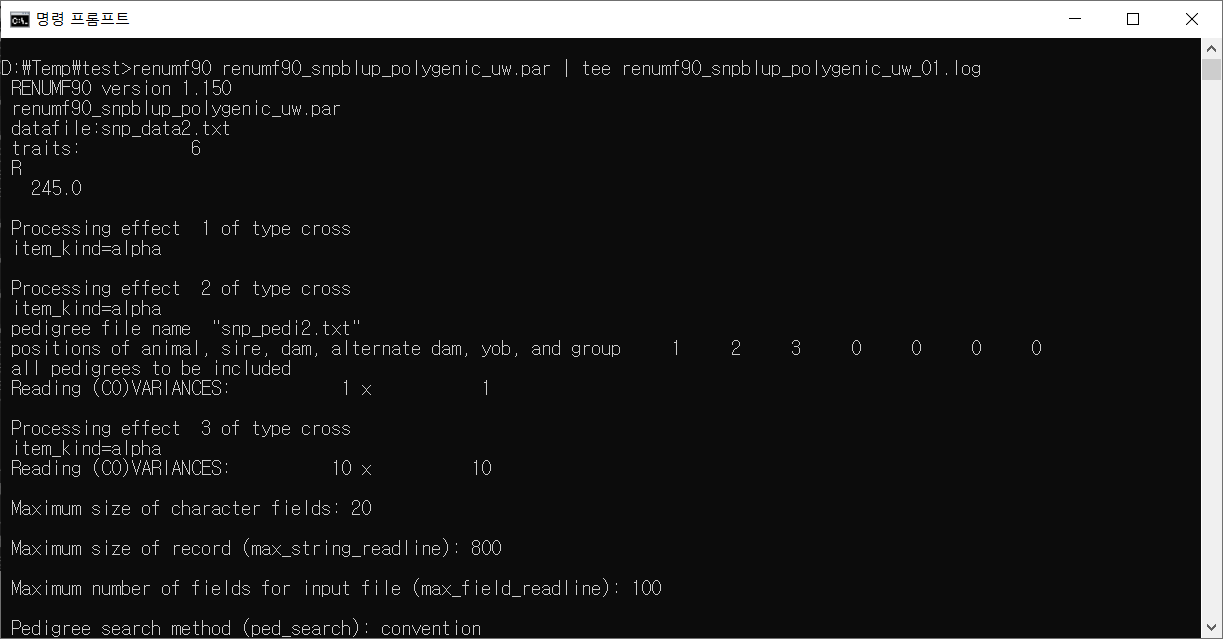
renumf90 실행 로그
RENUMF90 version 1.150
renumf90_snpblup_polygenic_uw.par
datafile:snp_data2.txt
traits: 6
R
245.0
Processing effect 1 of type cross
item_kind=alpha
Processing effect 2 of type cross
item_kind=alpha
pedigree file name "snp_pedi2.txt"
positions of animal, sire, dam, alternate dam, yob, and group 1 2 3 0 0 0 0
all pedigrees to be included
Reading (CO)VARIANCES: 1 x 1
Processing effect 3 of type cross
item_kind=alpha
Reading (CO)VARIANCES: 10 x 10
Maximum size of character fields: 20
Maximum size of record (max_string_readline): 800
Maximum number of fields for input file (max_field_readline): 100
Pedigree search method (ped_search): convention
Order of pedigree animals (animal_order): default
Order of UPG (upg_order): default
Missing observation code (missing): 0
hash tables for effects set up
first 3 lines of the data file (up to 70 characters)
13 0 0 1 558 9 0.00179211 1.3571429 -0.3571429 0.2857143 0.2857143 -0.
14 0 0 1 722 13.4 0.00138504 0.3571429 -0.3571429 -0.7142857 -0.714285
15 13 4 1 300 12.7 0.00333333 0.3571429 0.6428571 1.2857143 0.2857143
read 8 records
table with 1 elements sorted
added count
Effect group 1 of column 1 with 1 levels
table expanded from 10000 to 10000 records
added count
Effect group 2 of column 1 with 8 levels
table with 1 elements sorted
added count
Effect group 3 of column 1 with 1 levels
table expanded from 10000 to 10000 records
wrote statistics in file "renf90.tables"
Basic statistics for input data (missing value code is '0')
Pos Min Max Mean SD N
6 4.8000 15.400 9.8875 3.7434 8
random effect 2
type:animal
opened output pedigree file "renadd02.ped"
read 26 pedigree records
loaded 18 parent(s) in round 0
Pedigree checks
Number of animals with records = 8
Number of parents without records = 18
Total number of animals = 26
random effect 3
type:diag
Wrote parameter file "renf90.par"
Wrote renumbered data "renf90.dat" 8 records
Wrote field information "renf90.fields" for 14 fields in data
renumf90 실행으로 생긴 파일
renadd02.ped
26 3 20 1 0 2 0 0 0 26
1 0 0 3 0 0 1 1 0 13
21 9 11 1 0 2 0 0 0 21
13 0 0 3 0 0 0 0 1 5
2 3 17 1 0 2 1 0 0 19
3 0 0 3 0 0 1 8 0 14
22 3 16 1 0 2 0 0 0 22
11 0 0 3 0 0 0 0 1 3
16 0 0 3 0 0 0 0 1 8
4 1 12 1 0 2 1 2 0 15
23 3 19 1 0 2 0 0 0 23
18 0 0 3 0 0 0 0 1 10
9 0 0 3 0 0 0 1 0 1
14 0 0 3 0 0 0 0 1 6
5 4 10 1 0 2 1 0 0 16
24 3 18 1 0 2 0 0 0 24
19 0 0 3 0 0 0 0 1 11
12 0 0 3 0 0 0 0 1 4
6 4 13 1 0 2 1 0 0 17
17 0 0 3 0 0 0 0 2 9
25 3 15 1 0 2 0 0 0 25
20 0 0 3 0 0 0 0 1 12
7 3 17 1 0 2 1 0 0 20
10 0 0 3 0 0 0 0 1 2
8 3 14 1 0 2 1 0 0 18
15 0 0 3 0 0 0 0 1 7
renf90.dat
9 1.3571429 -0.3571429 0.2857143 0.2857143 -0.2857143 -1.2142857 -0.1428571 0.07142857 -0.1428571 1.2142857 1 1 1
13.4 0.3571429 -0.3571429 -0.7142857 -0.7142857 -0.2857143 0.7857143 -0.1428571 0.07142857 -0.1428571 -0.7857143 1 3 1
12.7 0.3571429 0.6428571 1.2857143 0.2857143 0.7142857 -1.2142857 -0.1428571 0.07142857 -0.1428571 1.2142857 1 4 1
15.4 -0.6428571 -0.3571429 1.2857143 0.2857143 -0.2857143 -0.2142857 -0.1428571 0.07142857 0.8571429 0.2142857 1 5 1
5.9 -0.6428571 0.6428571 0.2857143 1.2857143 -0.2857143 -1.2142857 -0.1428571 0.07142857 -0.1428571 1.2142857 1 6 1
7.7 0.3571429 0.6428571 -0.7142857 0.2857143 -0.2857143 0.7857143 -0.1428571 0.07142857 0.8571429 0.2142857 1 8 1
10.2 -0.6428571 -0.3571429 0.2857143 0.2857143 -0.2857143 0.7857143 -0.1428571 0.07142857 0.8571429 -0.7857143 1 2 1
4.8 -0.6428571 0.6428571 0.2857143 -0.7142857 -0.2857143 -0.2142857 -0.1428571 0.07142857 0.8571429 -0.7857143 1 7 1
renf90.par
# BLUPF90 parameter file created by RENUMF90
DATAFILE
renf90.dat
NUMBER_OF_TRAITS
1
NUMBER_OF_EFFECTS
12
OBSERVATION(S)
1
WEIGHT(S)
EFFECTS: POSITIONS_IN_DATAFILE NUMBER_OF_LEVELS TYPE_OF_EFFECT[EFFECT NESTED]
12 1 cross
13 26 cross
2 1 cov 14
3 1 cov 14
4 1 cov 14
5 1 cov 14
6 1 cov 14
7 1 cov 14
8 1 cov 14
9 1 cov 14
10 1 cov 14
11 1 cov 14
RANDOM_RESIDUAL VALUES
245.00
RANDOM_GROUP
2
RANDOM_TYPE
add_animal
FILE
renadd02.ped
(CO)VARIANCES
3.5241
RANDOM_GROUP
3 4 5 6 7 8 9 10 11 12
RANDOM_TYPE
diagonal
FILE
(CO)VARIANCES
8.9640 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000
0.0000 8.9640 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000
0.0000 0.0000 8.9640 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 8.9640 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 8.9640 0.0000 0.0000
0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 8.9640 0.0000
0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 8.9640
0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
8.9640 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 8.9640 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 8.9640
OPTION solv_method FSPAK
renf90.fields
field variable origfield group column random effect file
1 trait 6 0 0 * cov *
2 covariate 8 0 0 * cov *
3 covariate 9 0 0 * cov *
4 covariate 10 0 0 * cov *
5 covariate 11 0 0 * cov *
6 covariate 12 0 0 * cov *
7 covariate 13 0 0 * cov *
8 covariate 14 0 0 * cov *
9 covariate 15 0 0 * cov *
10 covariate 16 0 0 * cov *
11 covariate 17 0 0 * cov *
12 renumbered 4 1 1 * cross *
13 renumbered 1 2 1 animal cross renadd02.ped
14 renumbered 4 3 1 diagonal cross *
blupf90 실행 화면
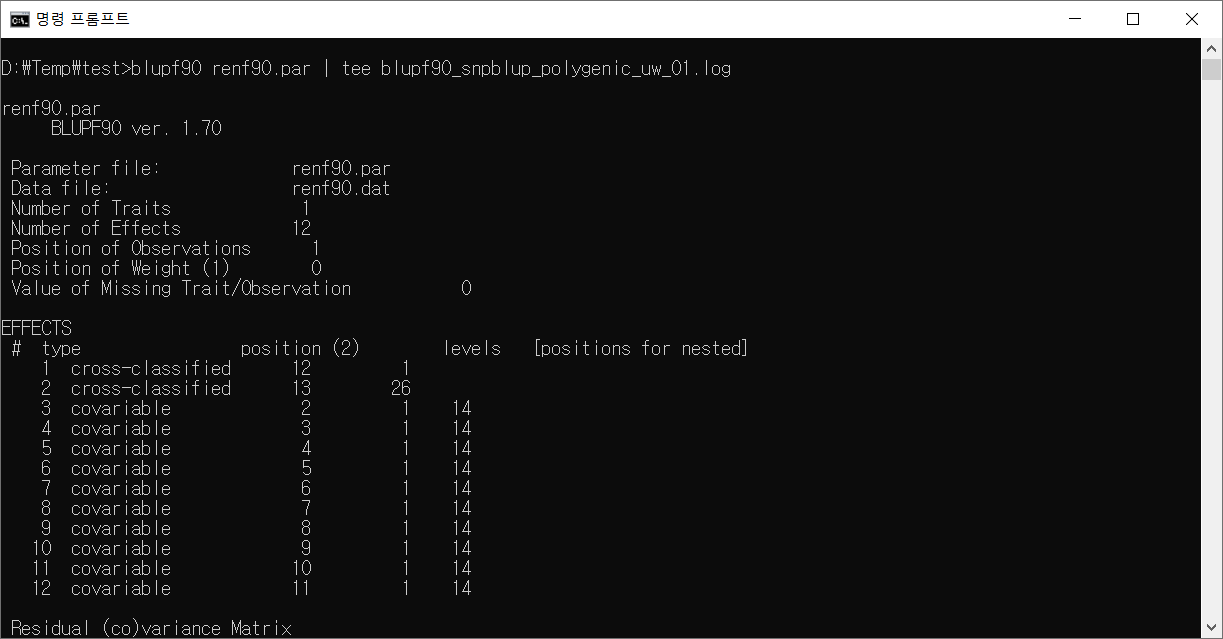
blupf90 실행 로그
renf90.par
BLUPF90 ver. 1.70
Parameter file: renf90.par
Data file: renf90.dat
Number of Traits 1
Number of Effects 12
Position of Observations 1
Position of Weight (1) 0
Value of Missing Trait/Observation 0
EFFECTS
# type position (2) levels [positions for nested]
1 cross-classified 12 1
2 cross-classified 13 26
3 covariable 2 1 14
4 covariable 3 1 14
5 covariable 4 1 14
6 covariable 5 1 14
7 covariable 6 1 14
8 covariable 7 1 14
9 covariable 8 1 14
10 covariable 9 1 14
11 covariable 10 1 14
12 covariable 11 1 14
Residual (co)variance Matrix
245.00
Random Effect(s) 2
Type of Random Effect: additive animal
Pedigree File: renadd02.ped
trait effect (CO)VARIANCES
1 2 3.524
correlated random effects 3 4 5 6 7 8 9 10 11 12
Type of Random Effect: diagonal
trait effect (CO)VARIANCES
1 3 8.964 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000
1 4 0.000 8.964 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000
1 5 0.000 0.000 8.964 0.000 0.000 0.000 0.000 0.000 0.000 0.000
1 6 0.000 0.000 0.000 8.964 0.000 0.000 0.000 0.000 0.000 0.000
1 7 0.000 0.000 0.000 0.000 8.964 0.000 0.000 0.000 0.000 0.000
1 8 0.000 0.000 0.000 0.000 0.000 8.964 0.000 0.000 0.000 0.000
1 9 0.000 0.000 0.000 0.000 0.000 0.000 8.964 0.000 0.000 0.000
1 10 0.000 0.000 0.000 0.000 0.000 0.000 0.000 8.964 0.000 0.000
1 11 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 8.964 0.000
1 12 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.000 8.964
REMARKS
(1) Weight position 0 means no weights utilized
(2) Effect positions of 0 for some effects and traits means that such
effects are missing for specified traits
* The limited number of OpenMP threads = 4
* solving method (default=PCG):FSPAK
Data record length = 14
# equations = 37
G
3.5241
G
8.9640 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000
0.0000 8.9640 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000
0.0000 0.0000 8.9640 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000
0.0000 0.0000 0.0000 8.9640 0.0000 0.0000 0.0000 0.0000 0.0000
0.0000
0.0000 0.0000 0.0000 0.0000 8.9640 0.0000 0.0000 0.0000 0.0000
0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 8.9640 0.0000 0.0000 0.0000
0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 8.9640 0.0000 0.0000
0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 8.9640 0.0000
0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 8.9640
0.0000
0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
8.9640
read 8 records in 0.2187500 s, 162
nonzeroes
read 26 additive pedigrees
finished peds in 0.2187500 s, 215 nonzeroes
solutions stored in file: "solutions"
blupf90 실행 결과 : solutions
trait/effect level solution
1 1 1 9.93695036
1 2 1 0.01067048
1 2 2 -0.01577057
1 2 3 0.00049787
1 2 4 0.04298274
1 2 5 0.07718088
1 2 6 -0.01695885
1 2 7 -0.05195194
1 2 8 -0.02022509
1 2 9 0.00000000
1 2 10 0.03712634
1 2 11 0.00000000
1 2 12 0.02509833
1 2 13 -0.02563348
1 2 14 -0.01364935
1 2 15 -0.00000000
1 2 16 -0.00000000
1 2 17 -0.03411019
1 2 18 -0.00000000
1 2 19 -0.00000000
1 2 20 -0.00000000
1 2 21 0.00000000
1 2 22 0.00024894
1 2 23 0.00024894
1 2 24 0.00024894
1 2 25 0.00024894
1 2 26 0.00024894
1 3 1 0.07836105
1 4 1 -0.28018933
1 5 1 0.23400811
1 6 1 -0.07435738
1 7 1 0.09844808
1 8 1 0.12723456
1 9 1 0.00000000
1 10 1 0.00000000
1 11 1 -0.05409385
1 12 1 -0.01787671
첫째 효과는 general mean, 둘째 효과는 개체의 residual polygenic effects, 3 ~ 12까지의 효과는 10개의 snp effect이다.
snpblup_polygenic_uw_blupf90.zip
0.00MB